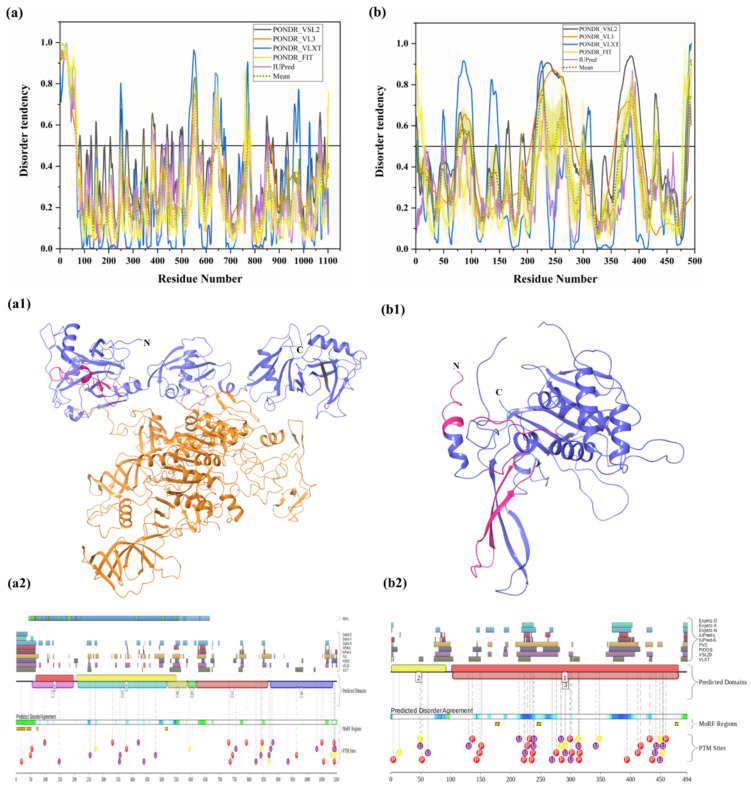
Figure 8.
Intrinsic disorder predisposition and structural characterization of human USP7 and USP14. (a) Disorder profile of human USP7 (UniProt ID: Q93009). (b) Disorder profile of human USP14 (UniProt ID: P54578) (a1) crystal structure of USP7 (PDB ID: 4YOC). (b1) Crystal structure of USP14 (PDB ID: 4GJQ). In Plots (a) and (b), disorder profiles generated by sets of disorder predictors such as PONDR® VSL2, PONDR® VL3, PONDR® VLXT, PONDR® FIT, and IUPred are depicted by black, orange, blue, yellow, and purple curves respectively. The mean disorder profile calculated from the average of five predictor-specific per-residue disorder profiles is shown by the olive color curve. Predicted disorder scores above 0.5 are considered disordered residues/regions. The light-olive shadow around mean curve represents the error distribution for the mean. The light-yellow shadow around the PONDR® FIT curve shows the error distribution for PONDR® FIT. In Plots a1 and b1, USP7 and USP14 are represented by faded blue color; in Plot (a1), human DNA (cytosine-5)-methyltransferase 1 (DNMT1) complexed with USP7 is shown in orange color. Disordered residues are shown by the salmon pink color. In Plot (a1), the positions of MoRFs (residues 1077–1082) predicted by the MoRFCHiBi_web server are shown by grey color in USP7 (PDB ID: 4YOC). (a2,b2) depict the PTM sites and MoRF regions obtained from the D2P2 server.