Figure EV5. Illustration of proposed model for the ability of Tat‐P4‐(C5)2 to increase macroscopic off‐rate of PICK1 bound to receptors embedded in the membrane.
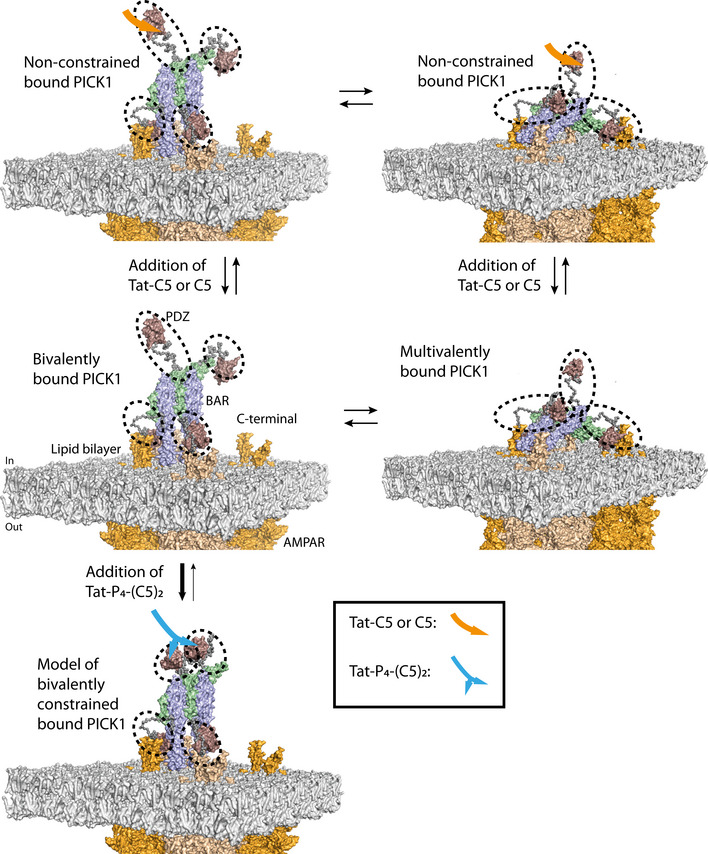
PICK1 binds to membrane‐embedded receptors, such as the AMPARs (orange), in two distinct states, a bivalent and a multivalent bound conformation, mainly driven by lipid binding motifs (black dashed circles) positioned in the PDZ domain and the amphipathic BAR‐PDZ linker (Erlendsson et al, 2019). Upon addition of Tat‐P4‐(C5)2, two or more PDZ domains of the PICK1 tetramer and the lipid binding motifs gets constrained, thereby favoring an upright position of PICK1, with a maximum of two PDZ domains and lipid binding regions of the PICK1 tetramer binding to the membrane and membrane‐embedded receptors. Addition of Tat‐C5 or C5 does not constrain the PDZ domains, which are then free to exchange between the bivalent upright PICK1 and the multivalent membrane parallel bound PICK1, thereby not affecting the macroscopic off‐rate. The model was created using PyMoL 2.0. The tetrameric PICK1 model was generated in earlier EOM modeling, and the position of the model is based on earlier work (Erlendsson et al, 2019). The structure of AMPARs (PDB: 5VHW) was positioned into a POPE lipid bilayer manually, and a de novo model of the GluA2 cytoplasmic tail (UniProt: P42262, residues 834‐883) was generated using PEP‐FOLD 3 (Thevenet et al, 2012; Shen et al, 2014; Lamiable et al, 2016) and orientated to the C‐terminal helix of the AMPAR structure manually.
