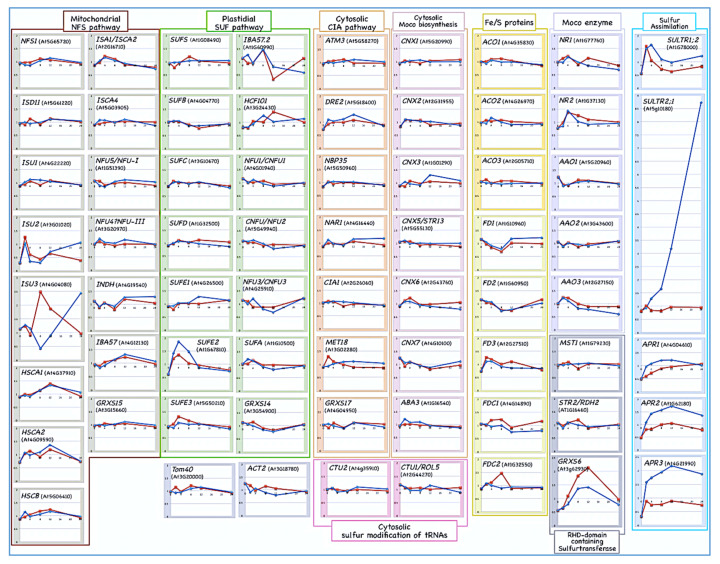
Figure 4.
Arabidopsis plants of 10-day-old grown on sulfate-containing medium were transferred to the medium with (the same concentration of) or without sulfate, and time course values of the transcripts were monitored for 24 h after sulfate repletion or depletion from the medium. Transcript levels of the genes related to the mitochondrial NFS, plastidial SUF, and cytosolic CIA pathways for Fe/S cluster biosynthesis and to some Fe/S proteins (aconitases and ferredoxins) are indicated. Gene expression levels of the transcripts of the cytosolic sulfur transfer pathways for Moco biosynthesis and sulfur modification of tRNAs also shown. Gene expression levels for some Moco-enzymes (two nitrate reductases NR1 and NR2, and three aldehyde oxidases AAO1, AAO2, and AAO3), some RHD-domain containing proteins such as mercaptopyruvate sulfurtransferases (MST1 and STR2/RDH2) and a cytosolic glutaredoxin, GRXS6, are also indicated. Transcript levels of several SULTRs and ARPs all of which are involved in the sulfur assimilation process are indicated as the highly responsive ones to the sulfate depletion [80]. Transcript levels of both TOM40 and ACT2 are also indicated because they are not related to the sulfur transfer systems mentioned here. All the microarray data used here are described in the previous paper [80]. Transcript levels were analyzed in sulfur-repleted (red line) or in sulfated-depleted (blue line) conditions. For each gene investigated here, gene name, and AGI code numbers are indicated on the top of the column.