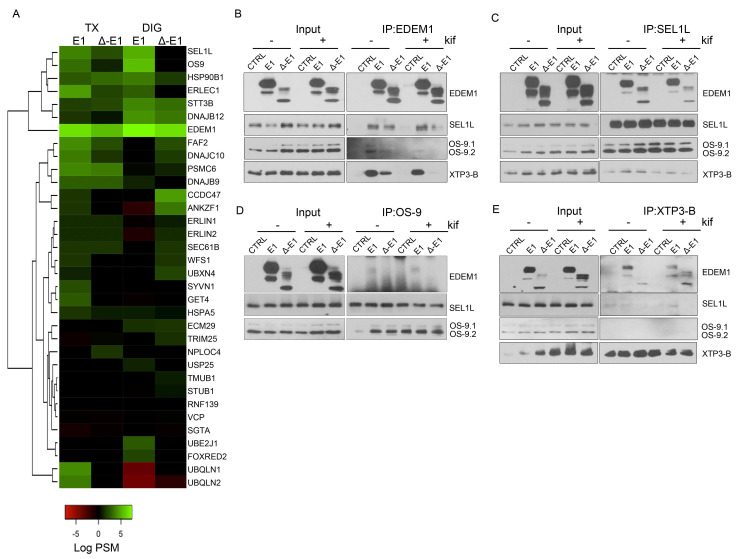
Figure 3.
Association of EDEM1 and ∆-EDEM1 with ERAD partner proteins. (A). Heatmap representation of proteins identified by mass spectrometry after enrichment with anti-EDEM1 antibodies from HEK293T cells overexpressing EDEM1 or ∆-EDEM1 extracted in Triton X-100 (TX) or digitonin (DIG)-containing buffers. The identified proteins were annotated with GO terms from UniProt database, and only entries that contained the ERAD key-term were further kept for analysis. The colour key denotes spectral counts (SC) in log scale after negative control (cells transfected with an empty vector and immunoprecipitated with EDEM1 antibodies) subtraction. (B–E). HEK293T cells were transfected to overexpress an empty vector (CTRL), EDEM1 (E1), and ∆-EDEM1 (∆-E1), and they were treated or not with 30 µM kifunensine (kif) ON. Cells were harvested, lysed in CHAPS-containing buffer, and equal amounts of protein were used for immunoprecipitation with antibodies for: (B) EDEM1, (C) SEL1L, (D) OS-9, and (E) XTP3-B. The eluted resin-bound complexes were separated by SDS-PAGE, and the membranes were probed with antibodies for EDEM1, SEL1L, OS-9, and XTP3-B.