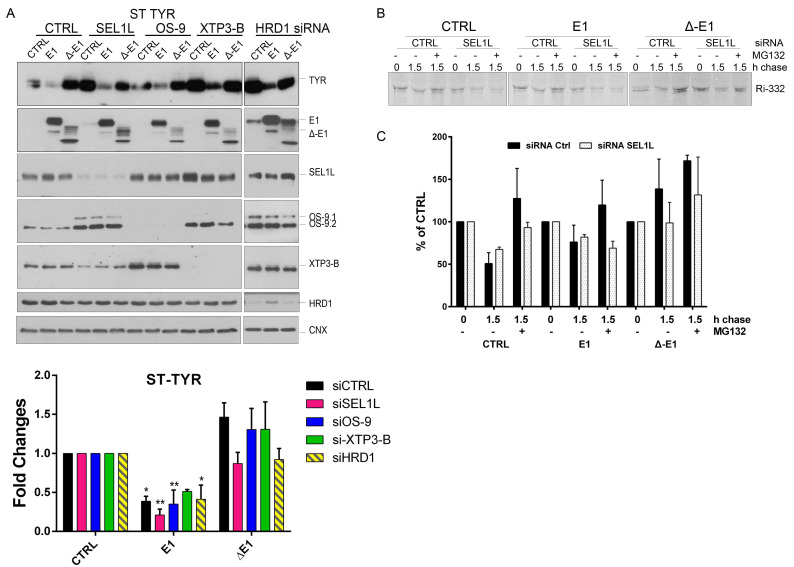
Figure A2.
(A) HEK293T cells were transfected with siRNA targeting a non-specific sequence (CTRL), SEL1L, OS-9, XTP3-B and HRD1 for 72 h; during the last 24 h another transfection was made to overexpress an empty vector (CTRL), EDEM1 (E1), and mutant ∆-EDEM1 (∆-E1) along with ST-Tyr. Cells were harvested and lysed, and the expression of Tyr, EDEM1, SEL1L, OS-9, XTP3-B, HRD1, and CNX as an internal control was estimated by Western blotting (upper panel); the lower panel depicts band densitometry plots of ST-Tyr (mean n = 3 ± SEM), and a two-way ANOVA comparison with a Bonferroni correction was applied for statistical analysis (* p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.001). For simplicity of representation, only statistically significant samples are indicated. (B,C) HEK293T were transfected with siRNA-targeting SEL1L or a non-specific sequence (CTRL) for 72 h; for the last 24 h, the cells were transiently transfected to co-express an empty vector (CTRL), EDEM1 (E1), or Δ-EDEM1 (Δ-E1) along with Ri-332. Cells treated or not with MG132 were metabolically labelled with 35S-Met/Cys for 20 min and chased for the indicated time points. Cells were harvested lysed in a Tritonx-100-containing buffer, and cleared lysates were subjected to immunoprecipitation with antibodies for Ribophorin I. The eluted samples were separated by SDS-PAGE followed by autoradiography. The band intensity of 2 independent experiments was estimated by densitometry measurement (using ImageJ software) as represented in the lower panel of the figure (mean n = 2 ± SEM).