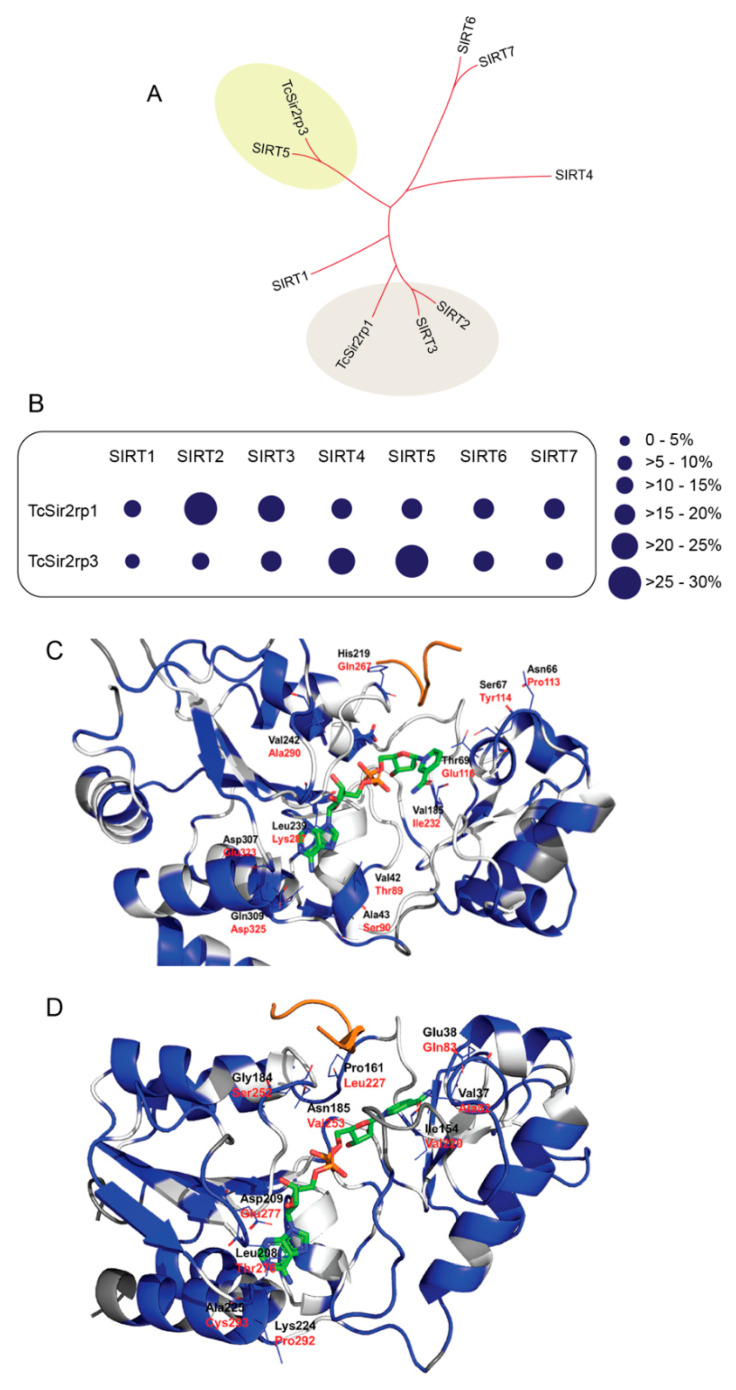
Figure 1.
Trypanosoma cruzi has divergent sirtuins. (A) Comparative phylogenetic analysis of T. cruzi and human sirtuins. (B) Schematic representation of amino acid identity of T. cruzi sirtuins compared to human sirtuins. (C) TcSir2rp1 homology model. Residues are colored by conservation with respect to human SIRT2 based on BLOSUM90 sequence alignment: white indicates conserved regions, blue indicates “mutated” regions and dark grey indicates not aligned regions. Residue names are indicated in black for TcSir2rp1 and red for SIRT2. NAD+ (green carbon atoms) and ALY (orange) were taken from 5Ol0 and 1ICI crystal structures respectively. (D) TcSir2rp3 homology model. Residues are colored by conservation with respect to human SIRT5 based on BLOSUM90 sequence alignment: White indicates conserved regions, blue indicates “mutated” regions and dark grey indicates not aligned regions. Residue names are indicated in black for TcSir2rp3 and red for SIRT5. NAD+ (green carbon atoms) and ALY (orange) were taken from 4I5I and 1S5P crystal structures respectively. HsSIRT1 (AAD40849.2); HsSIRT2 (AAD40850.2); HsSIRT3 (AAD40851.1); HsSIRT4 (AAD40852.1); HsSIRT5 (AAD40853.1); HsSIRT6 (NP_057623.2); and HsSIRT7 (NP_057622.1).