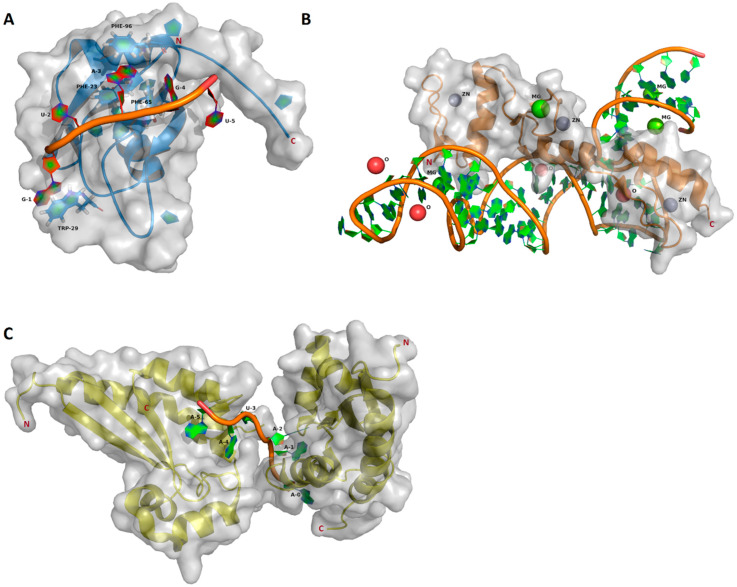
Figure 1.
Structures of the RNA binding domains, visualized with PyMOL version 1.5.0.4 using the corresponding RCSB protein data bank entries. Protein backbones are represented as blue (RNA recognition motif (RRM)), orange (Zinc finger domain (ZF)) and yellow (hnRNP-K homology (KH)) cartoon models. The solvent accessible surfaces are indicated in transparent gray. The bound RNA cartoon models are shown with filled aromatic rings. (A) Msi1 RRM1 in a complex with r(GUAGU) (NUMB Endocytic Adaptor Protein 5 (numb5)). The cartoon model contains filled aromatic rings. Protein side chains contributing to the RNA binding are shown as a stick model with filled aromatic rings (blue) (PDB ID: 2RS2) [14]. (B) ZF-RNA complex with ions. Solvent components are shown as balls: zink, grey; magnesium, green; oxygen (HOH), red (PDB ID: 1UN6) [42]. (C) Two KH domains in complex with r(AAAUAA). One KH domain binds the 5′AAA moiety and another KH binds the 3′UAA moiety (PDB ID: 5ELS) [43].