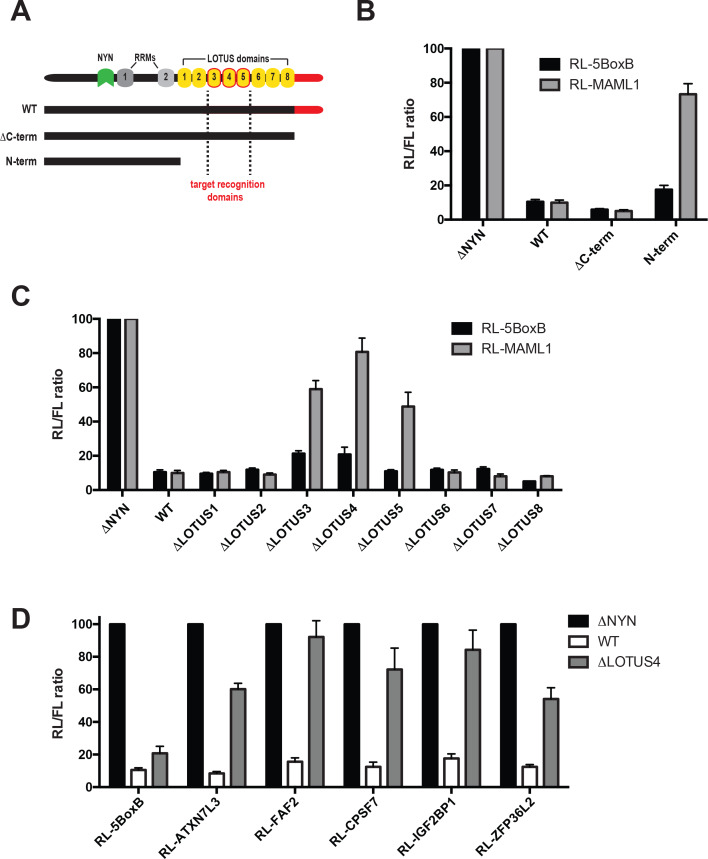
Figure 3. Central MARF1 LOTUS domains are required to silence mRNAs containing 3’UTRs of MARF1 target mRNAs.
(A) Schematic diagram of wild-type MARF1 protein, as well C-terminal deletion mutants. Core LOTUS domains required for MARF1-mediated repression are bordered in red. (B–D) RL activity detected in extracts from HEK293 cells expressing the indicated proteins. Cells were cotransfected with constructs expressing the RL-MAML1 reporter or RL-5BoxB reporter, along with FL, and indicated fusion proteins. Histograms represented normalized mean values of RL activity from a minimum of three experiments. RL activity values seen in the presence of λNHA-MARF1ΔNYN were set as 100.