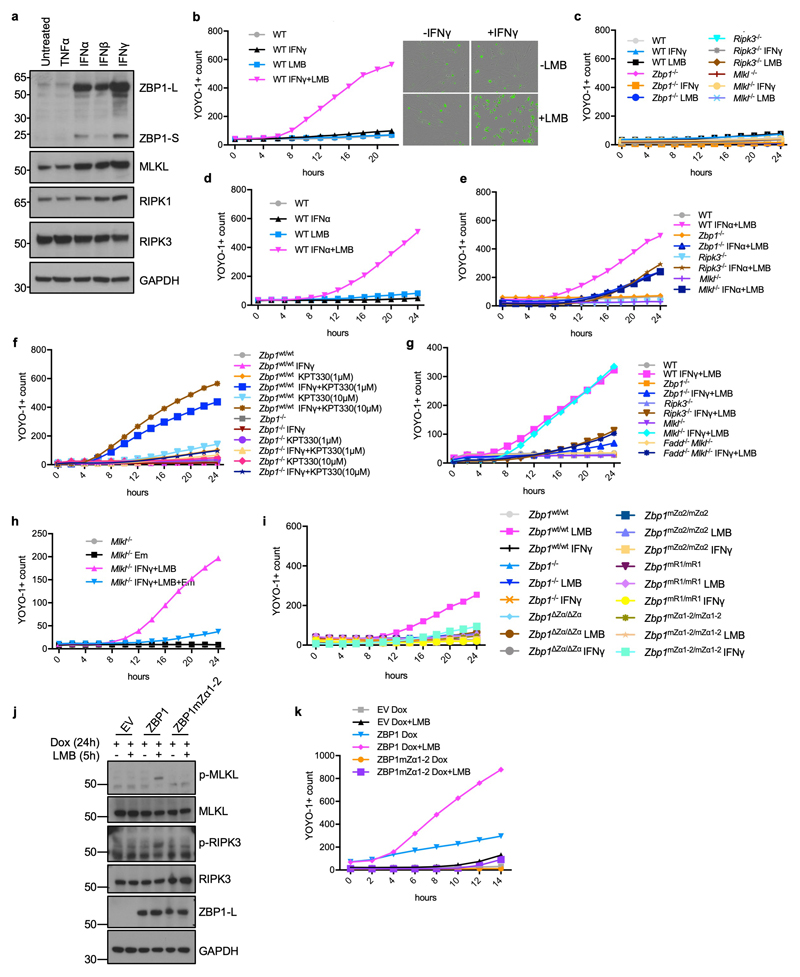
Extended Data Figure 9. Inhibition of nuclear export triggers Zα-dependent ZBP1-mediated cell death.
a, Immunoblot analysis of WT MEFs treated with TNFα (20 ng ml−1), IFNα (50 ng ml−1), IFNβ (50 ng ml−1) or IFNγ (1,000 u ml−1) for 24 h. b, Cell death measured by YOYO-1 uptake in WT MEFs treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment) and LMB (1 ng ml−1). IncuCyte images of WT MEFs before and after treatment with LMB for 24 h. YOYO-1 staining is shown in green. c-e, Graphs depicting cell death assessment by YOYO-1 uptake in MEFs with the indicated genotypes treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), IFNα (50 ng ml−1) (24 h pre-treatment) and LMB (1 ng ml−1). f, g, i, Graphs depicting cell death assessment by YOYO-1 uptake in LFs with the indicated genotypes treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), KPT-330 (1 μM or 10 μM) (f) and LMB (5 ng ml−1) (g, i). h, Graphs depicting cell death assessment by YOYO-1 uptake in Mlkl-/- MEFs treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), LMB (1 ng ml−1) and emricasan (5 μM). j, Immunoblot analysis of total lysates from iMEFs transduced with lentiviruses expressing Flag (EV), Flag-tagged ZBP1 or Flag-tagged ZBP1mZα1-2 stimulated with combinations of Dox (1 μg ml−1) (24 h pre-treatment) and LMB (5 ng ml−1). k, Graphs depicting cell death assessment by YOYO-1 uptake in iMEFs transduced with lentiviruses expressing Flag (EV), Flag-tagged ZBP1 or Flag-tagged ZBP1mZα1-2 stimulated with combinations of Dox (1 μg ml−1) (24 h pre-treatment) and LMB (5 ng ml−1). Dox, doxycycline. Representative data in panel a (n = 2), b (n = 6), c (n = 6), d (n = 6), e (n = 6), f (n = 4), g (n = 4), h (n = 4), i (n = 5), j (n = 2) and k (n = 3). Panels (b-i, k) show mean values from technical triplicates (n = 3). Data shown in panels c and i serve as controls for the data shown in Fig. 4a and 4e respectively, and come from the same experiments. GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.