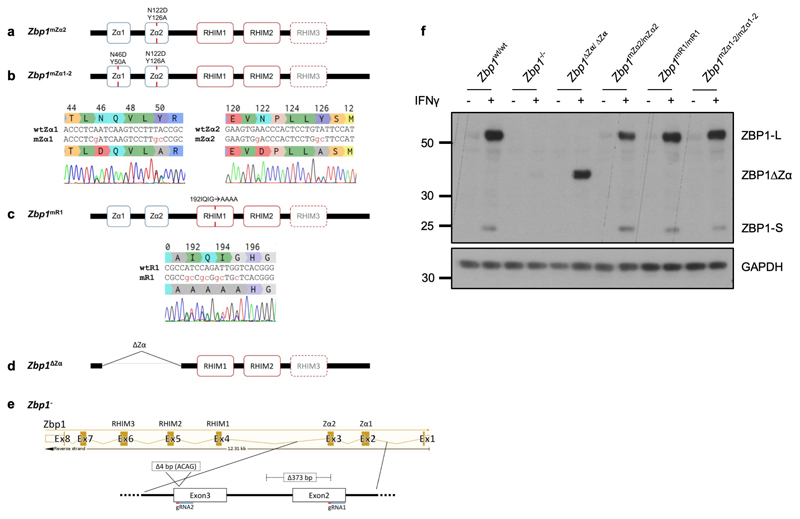
Extended Data Figure 1. Generation of mutant Zbp1 knock-in mice.
a-e, Schematic depicting the different Zbp1 mutant mice generated using CRISPR/Cas9-mediated gene targeting in C57BL/6N zygotes as indicated. To generate Zbp1mZα2 mice amino acids N122 and Y126 in Zα2 domain were substituted with D and A respectively (a). For Zbp1mZα1-2 mice both Zα1 (N46D and Y50A) and Zα2 (N122D, Y126A) were mutated (b). For Zbp1mR1 mice the residues of the core RHIM motif starting at amino acid position 192 (IQIG) were replaced by alanines (c). Sequencing traces of the desired mutations are shown in heterozygous mice. d, Zbp1ΔZα mice were generated by deleting exons two and three which contain both Zα domains (d). A new Zbp1- knockout mouse was generated by targeting both exons 2 and 3. An allele harboring a 373 bp deletion in exon 2 and the following intron as well as an additional 4 bp deletion in exon 3 was chosen as Zbp1 knockout allele (e). f, Immunoblot analysis of total lysates from LFs with the indicated genotypes stimulated with IFNγ (1,000 u ml−1) for 24 h. Data information: one representative out of two independent experiments shown (f). GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.