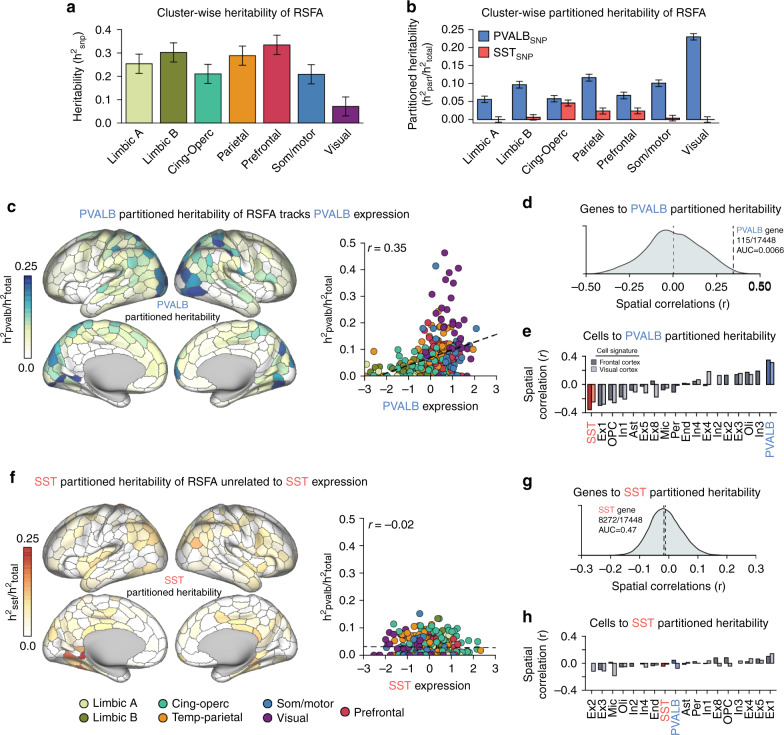
Fig. 5. PVALB-linked genetic variation explains patterns of heritable brain function.
a RSFA was significantly heritable across the seven empirically defined spatial clusters (n = 9713 UKB subjects). b Partitioned heritability analyses reveal that the 500-gene PVALBSNP set accounted for a significant proportion of heritable variance in all seven clusters (n = 9713 UKB subjects). c Parcel-wise PVALBSNP partitioned heritability of RSFA (left panel) tracks the PVALB single-gene expression across cortex (Pearson’s r(326) = 0.35, p = 1.04e−10; rs = 0.40, p = 2.2e−14). d Across all genes, PVALB was the 115th most correlated gene to PVALBSNP partitioned heritability (AUC = 0.007). e Across all deconvolved cell types, inferred PVALB cell fraction was the most positively correlated to PVALBSNP partitioned heritability (frontal cortex: Pearson’s r(329) = 0.35, p = 2.27e−11; rs = 0.39, p = 2.20e−13; visual cortex: Pearson’s r(329) = 0.31, p = 6.67e−9; rs = 0.34, p = 1.50e−10). f SSTSNP partitioned heritability was not associated with SST single-marker expression (Pearson’s r(326) = −0.02, p = 0.75), a null finding that was consistent when put in context of all genes (g) and inferred cell types (h). Error bars = standard error.