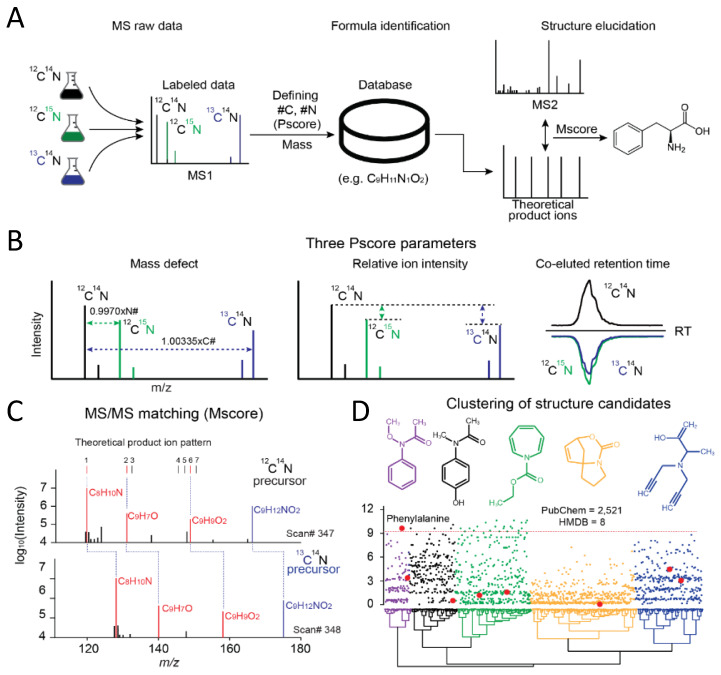
Figure 2.
JUMPm analysis of labeled samples and an example identification of phenylalanine. (A) Conceptual workflow for a stable isotope-labeled experiment (the double labeled sample is not shown); (B) The quality of each isotope-labeled pair is scored with three parameters (mass defect, relative ion intensity, and co-eluted retention time). The Pscore is used to discriminate authentic pairs from random matches; (C) For each isotope-labeled pair, the relevant MS2 spectra are scored (Mscore) and annotated with the top match. Three matched theoretical fragment ions are highlighted in red; (D) Hierarchical clustering of all structure candidates by predicted product ion intensities for the example metabolite spectrum (HMDB hits, large red dots and PubChem hits, small dots). Representative structures from each colored group are shown. All candidates share the neutral formula C9H11NO2.