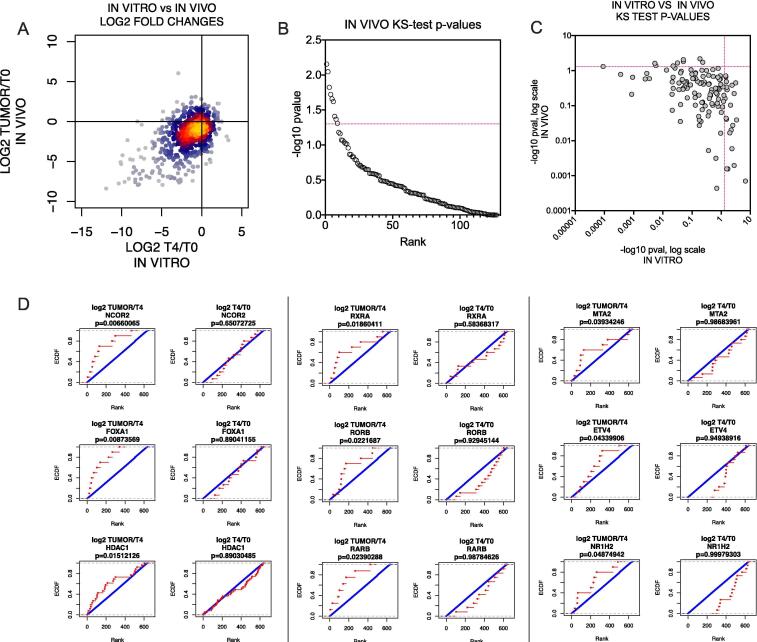
Fig. 2.
Identification of genes essential for in vivo tumorigenesis. (A) Comparison of shRNA abundance in the in vitro screen vs the in vivo screen. ShRNAs that were depleted in vivo but not in vitro are in the lower right quadrant. (B) Ranked plot of −log10p-values from a one-sided KS-test for each gene, comparing the distribution of log2 tumor/T4 fold changes in shRNAs against the gene to shRNAs of all other genes. Significance threshold (p ≤ 0.05; −log10 p ≥ 1.3) is indicated by a red dotted line. (C) Comparison of −log10p-values from KS-test for each gene in the in vitro screen (log2 T4/T0) vs the in vivo screen (log2 tumor/T4). Significance thresholds are indicated by red dotted lines (p ≤ 0.05; −log10p ≥ 1.3). Genes which scored as significantly depleted in vivo but not in vitro are represented in the upper left quadrant. (D) Cumulative distribution plots for the 9 screening hits which were significantly depleted in vivo but not in vitro. Left, in vivo plots; right, in vitro plots. The distribution of log2 fold changes of all shRNAs for that gene (red) are compared to the distribution of log2 fold changes of all other shRNAs. KS-test p-values are shown above each plot. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)