Figure 1.
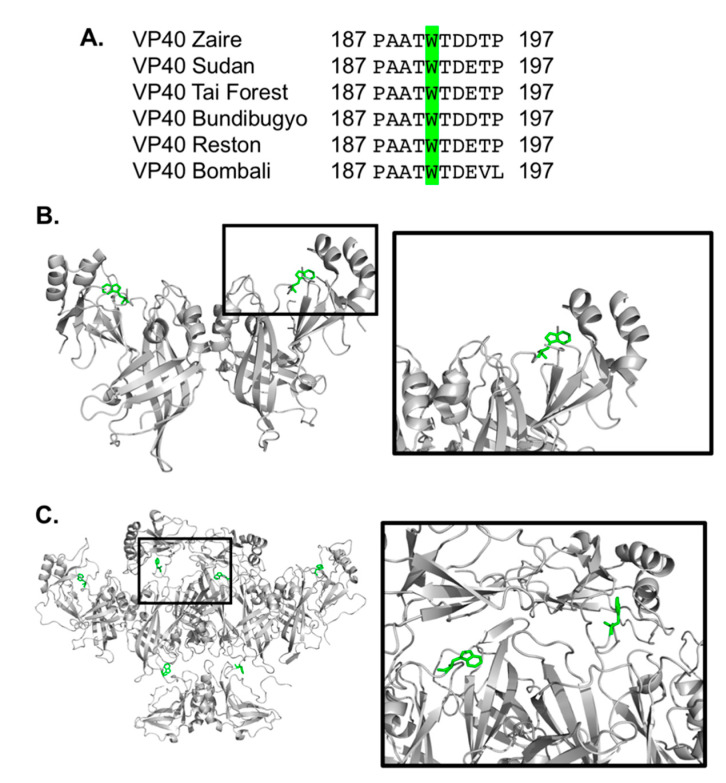
eVP40 has a conserved tryptophan at position 191. (A) Clustal W was used to align the amino acid sequences of eVP40 from six different types of ebolavirus. The alignment between residues 187 and 197 is shown with the conserved tryptophan residues (Trp191) highlighted in green. (B) The eVP40 dimer structure (PDB ID: 4LDB) was used to highlight the position of Trp191, which is situated near a C-terminal domain (CTD) region beneath Lys274 and Lys275, which are important for eVP40 plasma membrane binding [13,15]. (C) The VP40 hexamer structure (PDB ID: 4LDD) was used to model in the missing CTDs (Modeller software) to highlight the position of Trp191 in the proposed hexamer structure. Figure 1B,C were prepared in Pymol.