Figure 1. Challenge overview and initial conformations of the host-guest systems featured in the SAMPLing challenge.
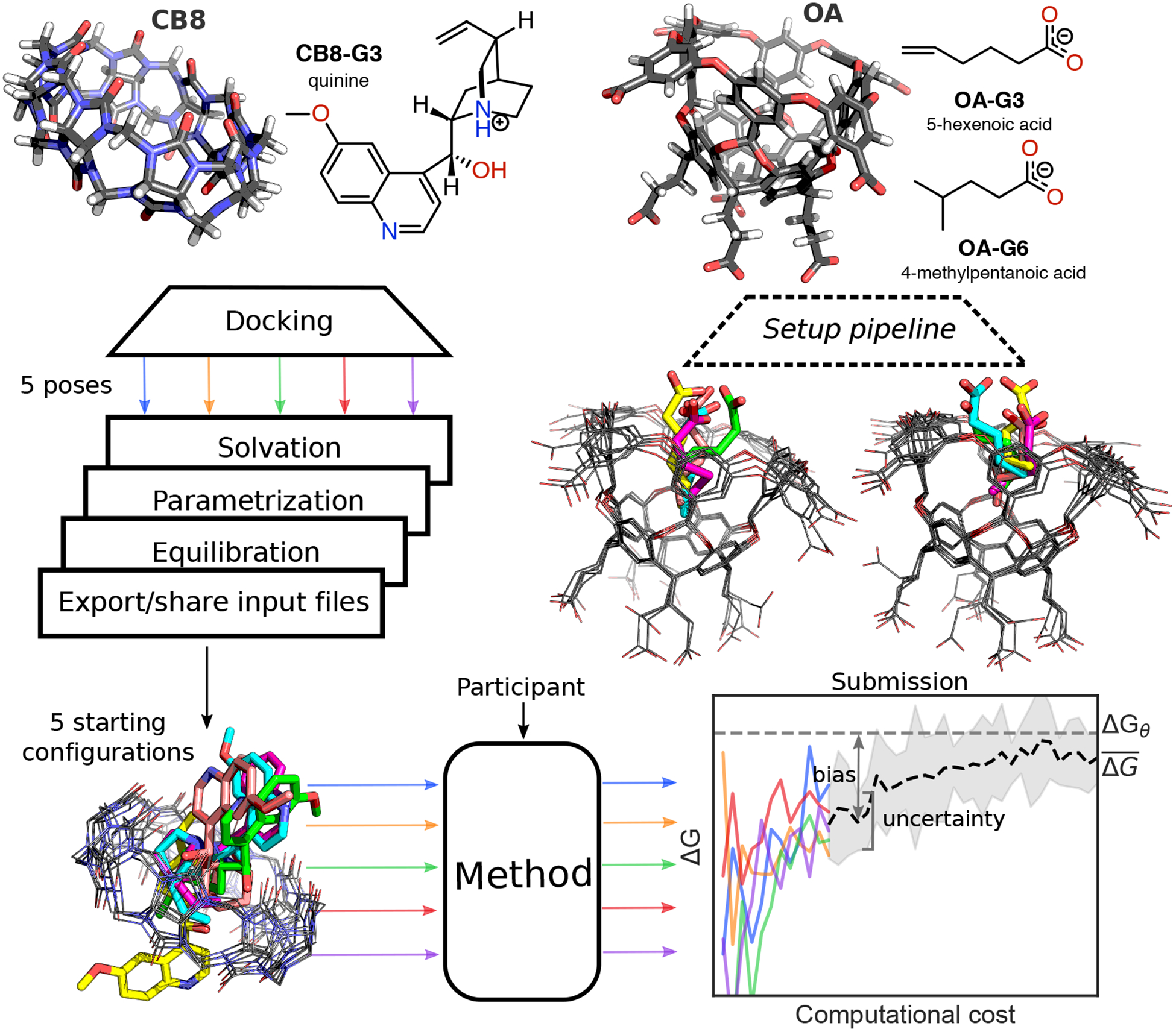
The three-dimensional structures of the two hosts (i.e. CB8 and OA) are shown with carbon atoms represented in black, oxygens in red, nitrogens in blue, and hydrogens in white. Both the two-dimensional chemical structures of the guest molecules and the three-dimensional structures of the hosts entering the SAMPLing challenge are shown in the protonation state used for the molecular simulations. We generated five different initial conformations for each of the three host-guest pairs through docking, followed by a short equilibration with Langevin dynamics. The three-dimensional structure overlays of the five conformations for CB8-G3, OA-G3, and OA-G6 are shown from left to right in the figure with the guests’ carbon atoms colored by conformation. Participants used the resulting input files to run their methods in five replicates and submitted the free energy trajectories as a function of the computational cost. We analyzed the submissions in terms of uncertainty of the mean binding free energy estimate and its bias with respect to the asymptotic free energy ΔGθ.
