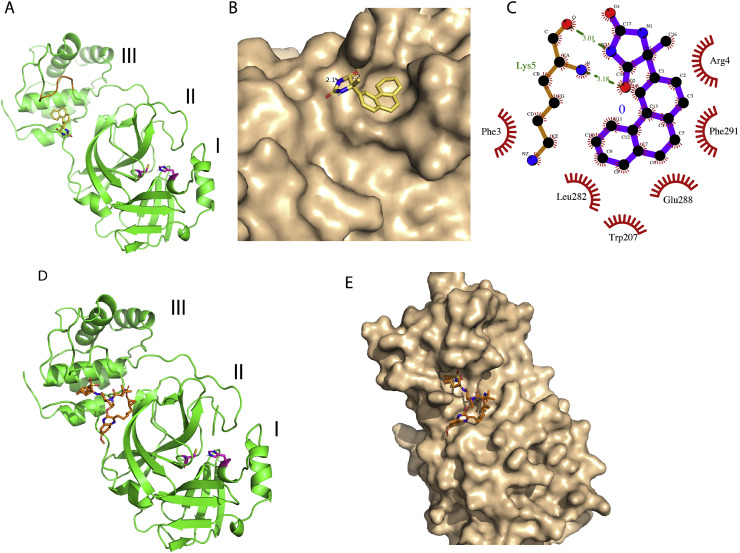
Fig. 2.
Representative low energy conformations of Bagrosin and Grazoprevir docked at an allosteric site of SARS-CoV-2 3CL-protease. (A) Cartoon presentation of SARS-CoV-2 3CL-protease monomer with individual domains labeled as I, II and III. The docked Bagrosin molecule is depicted in sticks with carbon, nitrogen and oxygen atoms colored yellow, blue and red, respectively. Two catalytic residues, His-41 and Cys-145 are also highlighted in purple sticks. The main region of domain III involved in forming the dimer interface is colored brown. (B) Molecular surface representation of 3CL-protease and bound Bagrosin depicted in sticks (C) Ligplot derived molecular interaction description of the docking complex, where H bonds are represented by dotted green lines. (D) and (E) Cartoon and surface representations of 3CL-protease, respectively including docked structure of Grazoprevir depicted in sticks.