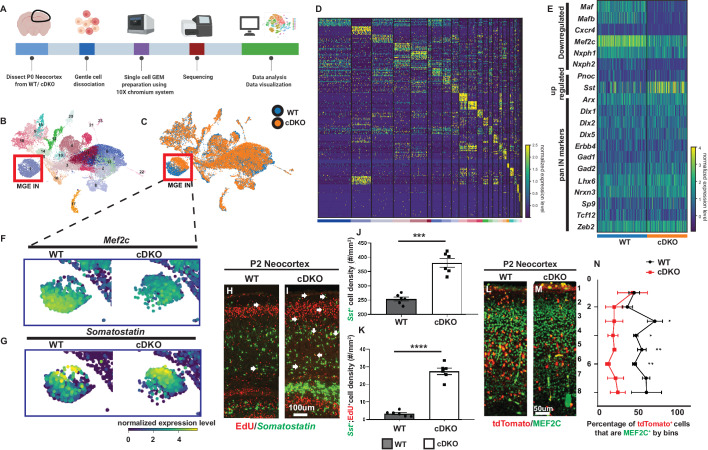
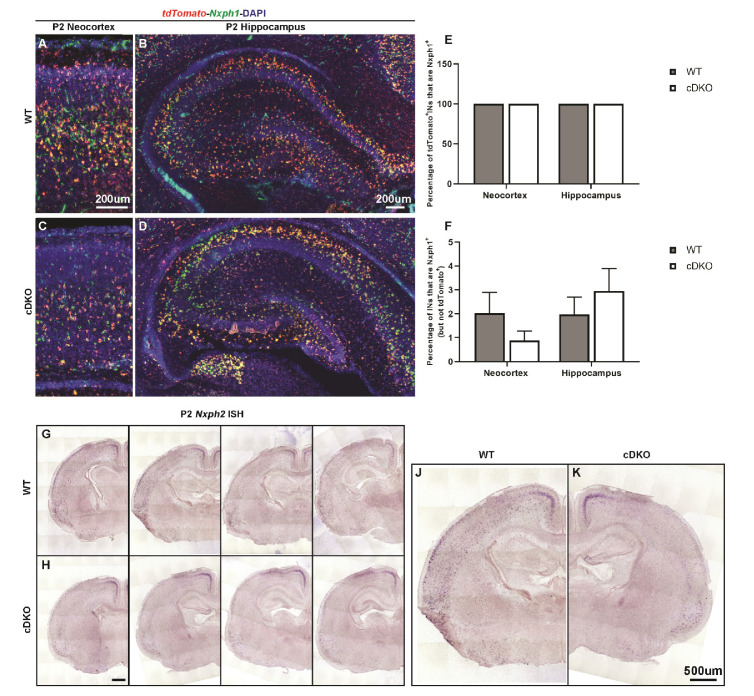
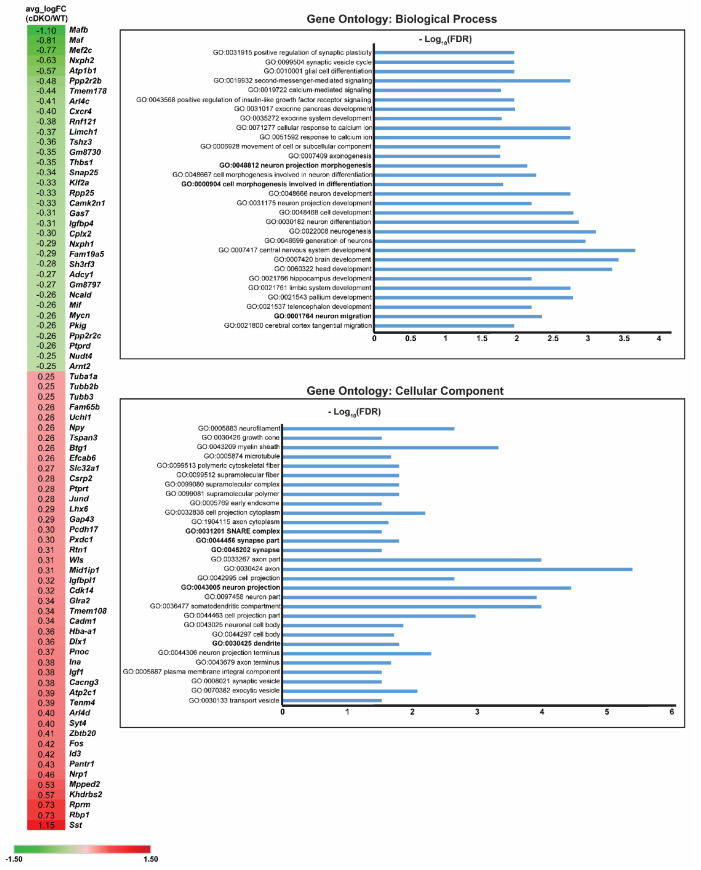
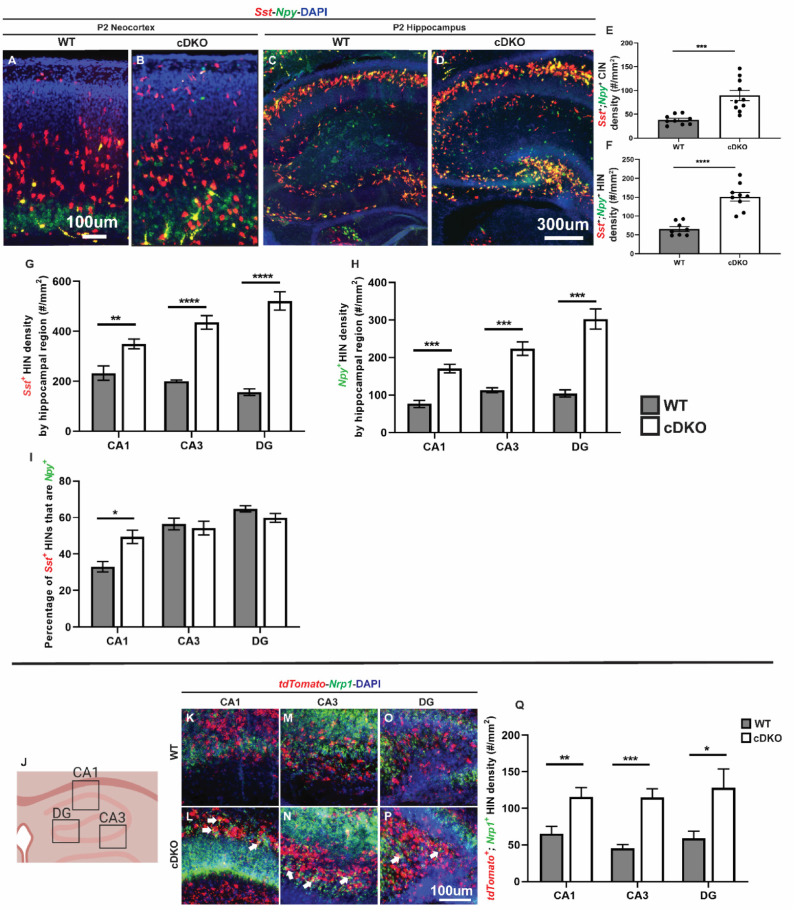
Figure 1. Single cell transcriptomic analysis of P0 WT and Maf cDKO neocortex.
(A) Experimental design for 10X genomics analysis of the P0 WT and Maf cDKO neocortex. Neocortical tissues were dissociated for single cell gel bead-in-emulsion (GEMs) particle preparation, followed by cDNA library preparation, sequencing and data analysis. (B) UMAP plot of WT and cDKO cells, color coded with cluster identities (see Figure 1—figure supplement 2). (C) UMAP plot of WT and cDKO cells, color coded with genotypes; cluster one are MGE INs. Note the genotype separation of the cells in cluster 1 (left bottom), but not other clusters, suggesting the lack of non-cell autonomous effects of the cDKO. (D) Heatmap showing cluster identities with corresponding marker genes. (E) Heatmap of cluster 1 cells from WT and cDKO showing average expression of selected MGE IN markers and pan-IN markers. Note the nearly full depletion of Mafb and Maf and increased expression of Sst, and the decreased expression of Mef2c, while other MGE-derived IN markers such as Dlx1/2/5, Gad2 and Lhx6 were not changed. This suggests that the deletion of Mafs in the MGE-lineage does not lead to a gross change in MGE-derived IN fate. (F–G) Enlarged feature plots showing the expression of Mef2c (F) and Sst (G) from WT and cDKO from cluster 1. (H–I) FISH in Sst together with EdU labeling showing the amount of E15.5 born MGE-derived CINs that became Sst+ in WTs and cDKOs. Arrows point to the cells that are EdU+;Sst+. (J) Quantification of Sst+ CINs at P2 WT and cDKO neocortex. (K) Quantification of EdU+;Sst+ CINs at P2 WT and cDKO neocortex. (L–M) Immunofluorescent staining of MEF2C colocalized with Nkx2.1-cre-driven tdTomato reporter. P2 Maf cDKOs have decreased MEF2C expression (M) compared to WTs (L). (N) Quantification of the proportion of tdTomato+ CINs that were MEF2C+ by bins. N = 3–4 animals per group and multiple brain sections were used for quantification. Scale bar in (I) = 100 um and in (M) = 50 um. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.