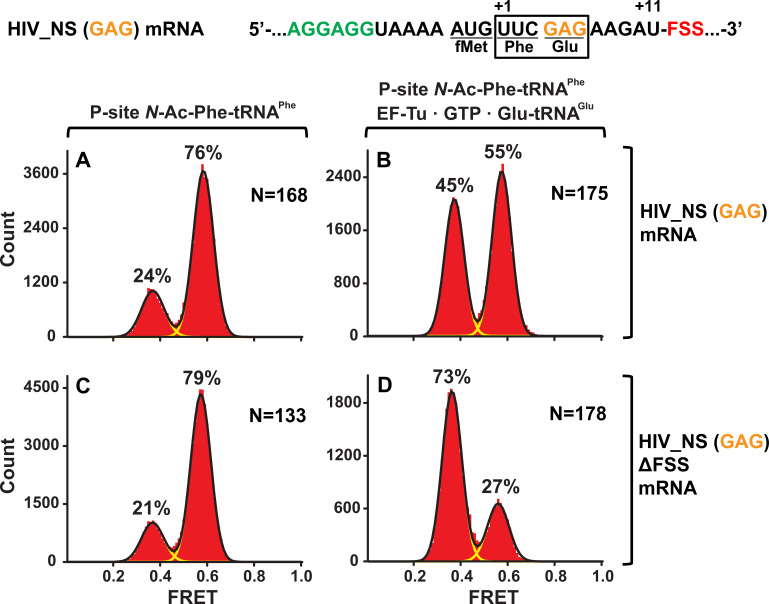
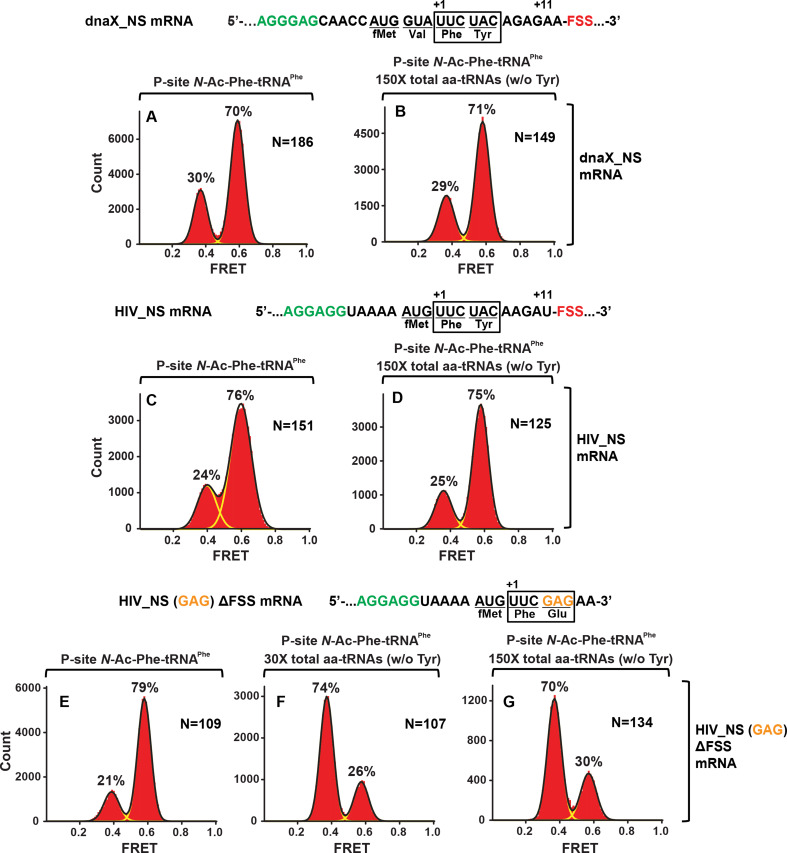
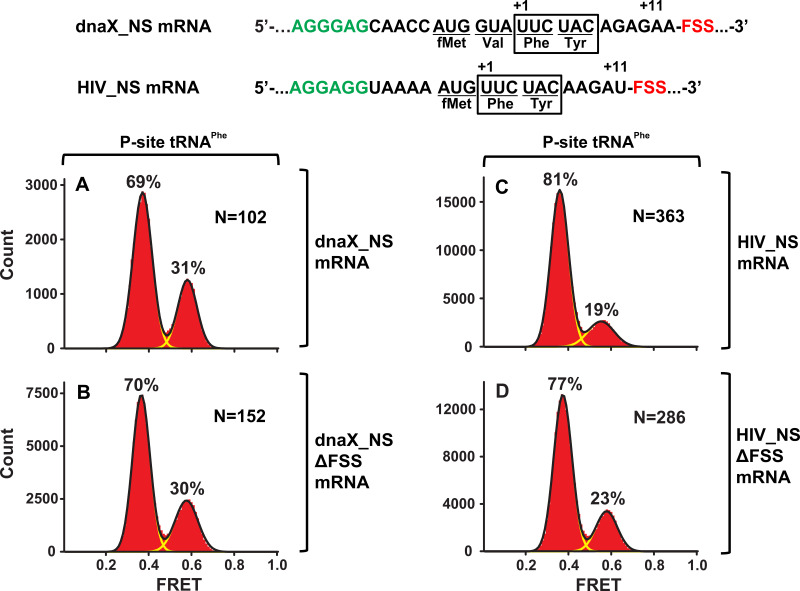
Figure 5. The FSS-induced ribosome stalling in NR conformation is independent of A-site codon identity.
Histograms show FRET distributions in S6-cy5/L9-cy3 ribosomes programmed with HIV_NS (GAG) (A–B) or HIV_NS (GAG) ∆FSS (C–D) mRNA, respectively. Ribosomes containing P-site N-Ac-Phe-tRNAPhe (A, C) were incubated with EF-Tu•GTP•Tyr-tRNATyr (B, D) for 5 min and imaged after removal of unbound aminoacyl-tRNAs. Yellow lines show individual Gaussian fits of FRET distributions. Black lines indicate the sum of Gaussian fits. N indicates the number of FRET traces compiled into each histogram. The fractions of the ribosome in R and NR conformations are shown above the corresponding 0.4 and 0.6 Gaussian peaks, respectively.