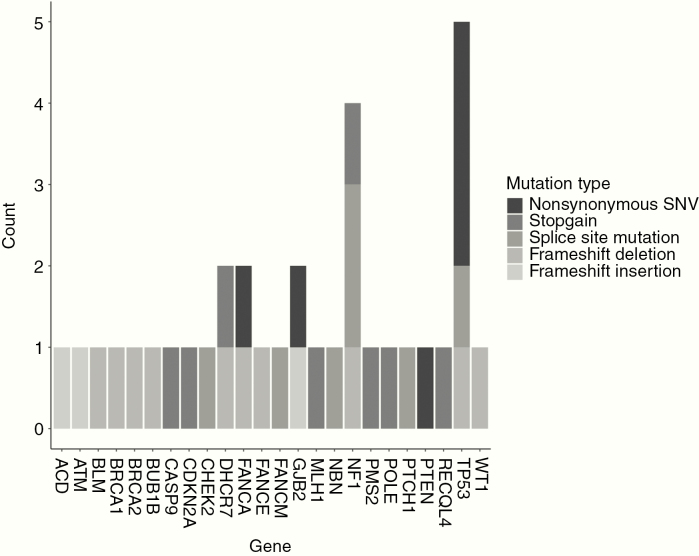
Fig. 2.
Prevalence of putatively pathogenic germline variants in cancer predisposition genes by variant type based on strict filtering criteria. Bars are shaded by variant type. Variants were deemed putatively pathogenic if deemed nonsense, frameshift, altering splice site, or annotated as “Pathogenic” or “Likely pathogenic” based on ClinVar and had an allele frequency ≤0.0001 in both TOPMed and gnomAD.