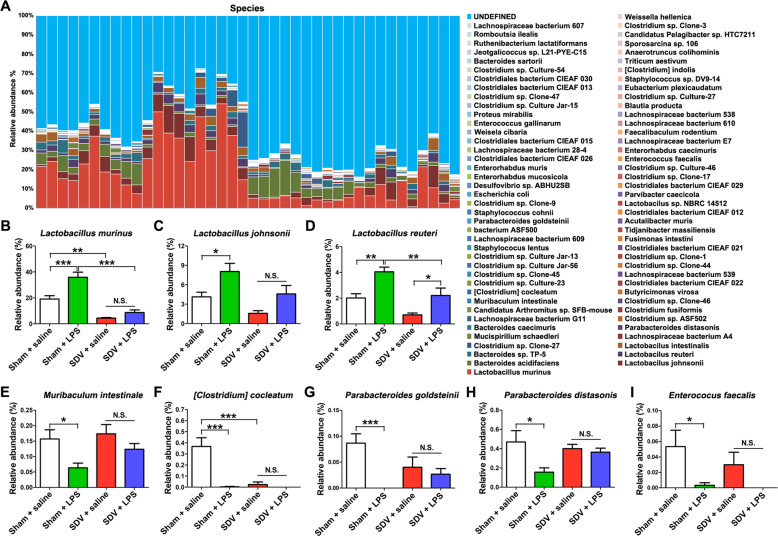
Fig. 6. Altered gut bacteria composition at the species level.
a Relative abundance at the species level in the four groups. bLactobacillus murinus (two-way ANOVA: LPS: F1,36 = 16.64, P = 0.0002; SDV: F1,36 = 66.09, P < 0.0001; interaction: F1,36 = 5.849, P = 0.02085). cLactobacillus johnsonii (two-way ANOVA: LPS: F1,36 = 12.13, P = 0.0013; SDV: F1,36 = 9.196, P = 0.0045; interaction: F1,36 = 0.212, P = 0.6479). dLactobacillus reuteri. (two-way ANOVA: LPS: F1,36 = 21.04, P < 0.0001; SDV: F1,36 = 16.75, P = 0.0012; interaction: F1,36 = 0.469, P = 0.4977). eMuribaculum intestinale (two-way ANOVA: LPS: F1,36 = 8.636, P = 0.0057; SDV: F1,36 = 2.471, P = 0.1247; interaction: F1,36 = 0.789, P = 0.3802). f[Clostridium] cocleatum (two-way ANOVA: LPS: F1,36 = 22.22, P < 0.0001; SDV: F1,36 = 17.86, P = 0.0002; interaction: F1,36 = 17.18, P = 0.0002). gParabacteroides goldsteinii (two-way ANOVA: LPS: F1,36 = 11.98, P = 0.0014; SDV: F1,36 = 0.479, P = 0.4932; interaction: F1,36 = 6.444, P = 0.0156). hParabacteroides distasonis (two-way ANOVA: LPS: F1,36 = 6.288, P = 0.0168; SDV: F1,36 = 0.959, P = 0.3340; interaction: F1,36 = 3.929, P = 0.0551). iEnterococcus faecalis (two-way ANOVA: LPS: F1,36 = 8.907, P = 0.0051; SDV: F1,36 = 0.990, P = 0.3265; interaction: F1,36 = 0.557, P = 0.4604). The data represent mean ± S.E.M. (n = 10). *P < 0.05, **P < 0.01, ***P < 0.0001; N.S. not significant.