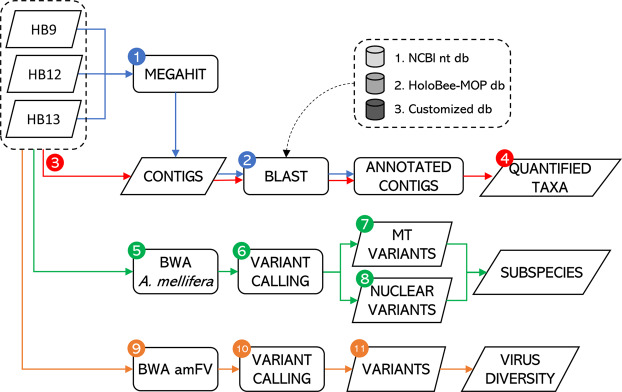
Figure 1.
Flowchart of the bioinformatic pipeline used to characterize the hologenome and environmental signature of the honey bee colony superorganism detected from honey DNA. Steps are as follows: (1) assembly of sequenced reads via MEGAHIT; (2) taxonomical assignment via BLASTN; (3) remapping of sequenced reads over the assembled contigs in order to (4) quantify the detected organisms; (5) mapping of sequenced reads via BWA over the Apis mellifera reference genome; (6) detection of genome variants; (7) subspecies identification through inspection of the mitochondrial DNA (mtDNA; method described by Utzeri et al.15; (8) subspecies identification via inspection of nuclear single nucleotide polymorphisms (SNPs; panel proposed by Muñoz et al.60); (9) mapping of sequenced reads via BWA over the Apis mellifera filamentous virus; (10) detection of genome variants; (11) estimation of virus diversity (Fixation index computation).