FIGURE 2.
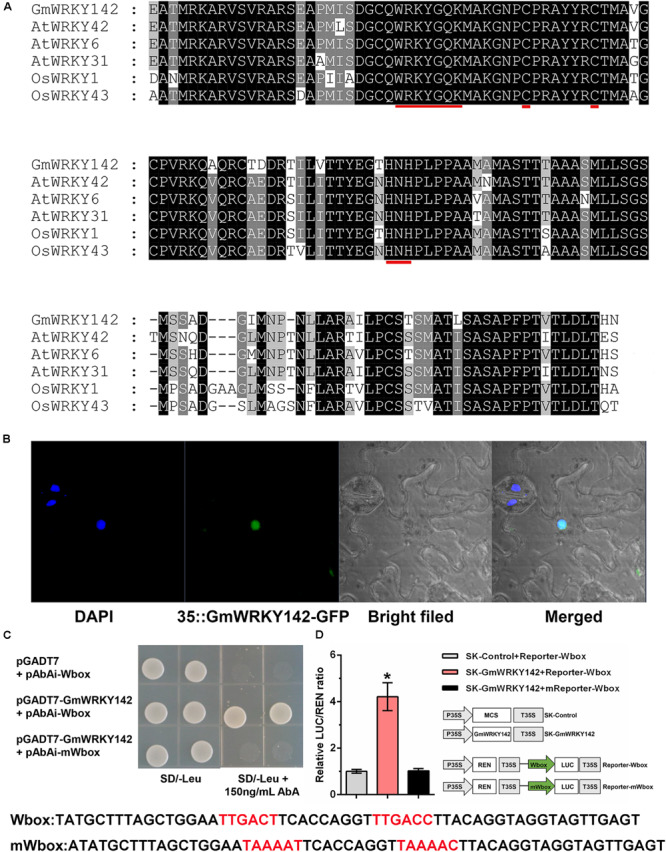
Sequence analysis, subcellular localization, and transcriptional activity assays of the GmWRKY142 protein. (A) Multiple alignments of the conserved WRKY domains between the WRKYs of soybean, rice and Arabidopsis. Identical amino acids are shown with a black background and analogous amino acids are shaded in gray. The WRKY motif and zinc-finger structures are denoted by red lines. (B) Subcellular localization of GmWRKY142 in Nicotiana benthamiana. DAPI was used to stain the nucleus. The overlapped images are shown on the right. (C) The growth phenotype of the cotransformant that harbored pGADT7-GmWRKY142 and bait vectors on a selective DO medium plate (SD/-Leu) with or without 150 ng mL–1 Aureobasidin A (AbA). (D) Schematic diagrams of the effector and reporter constructs used for transient luciferase (LUC) assays. Full-length CDS of GmWRKY142 was inserted into pGreen II 62-SK to produce an effector, while the original or mutated W-box elements were inserted into pGreen II 0800-LUC to generate reporters. MCS, multiple cloning site. P35S and T35S, the promoter and terminator of CaMV 35S, respectively. REN (Renilla luciferase) was used as an internal control for activity normalization. Transient expression assay of the promoter activity, shown as LUC/REN ratio, using tobacco leaves co-transformed with the effector and the reporters. LUC/REN ratio of the control co-transformed with the reporters and the empty effector vector (pGreen II 62-SK) was set as 1. Values are expressed as the means ± SD (n = 3). Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
