FIGURE 6.
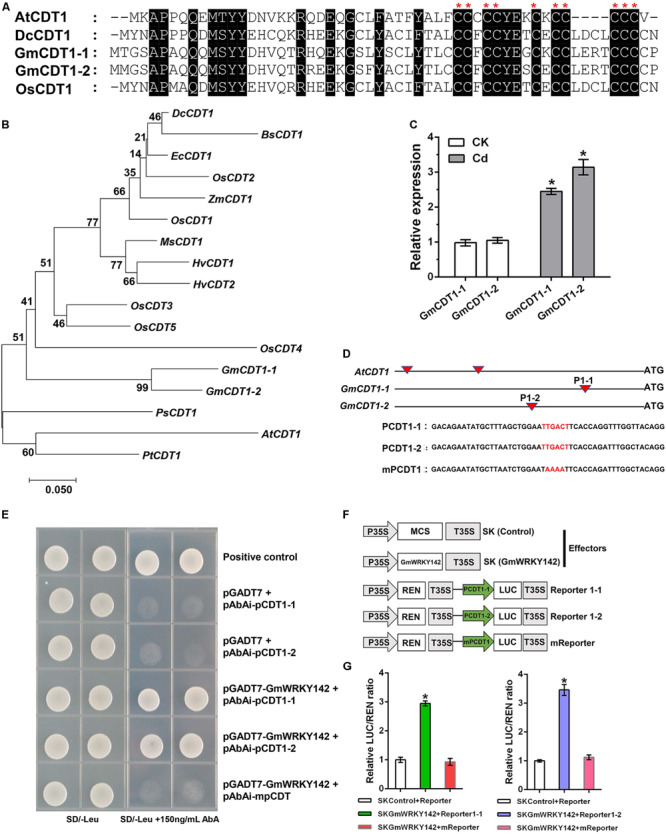
GmWRKY142 was bound to and activated the promoters of the GmCDT1-like genes. (A) Multiple alignments of the conserved CDT domains. Identical amino acids are shown with a black background. (B) Phylogenetic relationships between GmCDT1s and related proteins. Accession numbers are: AtCDT1, NM_202281; BsCDT1, ABL85053; EcCDT1, AB426477; HvCDT1, AK251605; HvCDT2, AK249080; MsCDT1, AB426478; OsCDT1, AK121052; OsCDT2, AK061597; OsCDT3, AK062450; OsCDT4, AK059641; OsCDT5, AK099514; PsCDT1, EF081525; PtCDT1, CU225257; ZmCDT1, AY103859. (C) GmCDT1-1 and GmCDT1-2 were induced by Cd. The Huaxia 7 roots cultured in 0 or 25 μM of CdCl2 for 6 h were sampled and qRT-PCR assay was performed. (D) Schematic diagrams of ATCDT1, GmCDT1-1, and GmCDT1-2 promoters and partial sequences containing W-box used in yeast one-hybrid assays. (E) Culture of yeast cells co-transformed with the prey and bait, as well as the positive control (pGADT7-Rec-p53+p53-AbAi) on selective medium with or without 150 ng mL–1 Aureobasidin A (AbA). (F) Schematic diagrams of the effector and reporter constructs used for transient luciferase (LUC) assays. Full-length CDS of GmWRKY142 was inserted into pGreen II 62-SK to produce an effector, while the original or mutated W-box elements were inserted into pGreen II 0800-LUC to generate reporters. MCS, multiple cloning site. P35S and T35S, the promoter and terminator of CaMV 35S, respectively. REN (Renilla luciferase) was used as an internal control for activity normalization. (G) Transient expression assay of the promoter activity, shown as LUC/REN ratio, using tobacco leaves co-transformed with the effector and the reporters. LUC/REN ratio of the control co-transformed with the reporters and the empty effector vector (pGreen II 62-SK) was set as 1. Values are expressed as the means ± SD (n = 3). The experiment was performed with at least three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
