FIGURE 4.
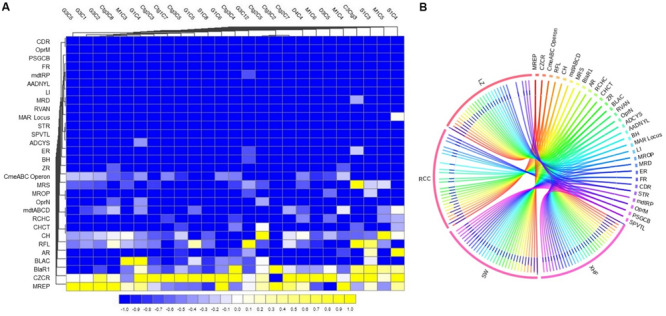
Projection of the resistance to antibiotic and toxic compounds (RATC) genes in bovine clinical mastitis (CM) pathogens. (A) Heatmap showing the hierarchical clustering of 30 different RATC genes detected in CM associated microbiomes of 25 CM milk samples as measured at level-3 of SEED subsystems in MG-RAST pipeline. The relative abundance of these genes significantly correlated (Pearson correlation, P = 0.002) with the relative abundance of the bacterial taxa found in these samples. The color bar at the bottom represents the relative abundance of putative genes and expressed as a value between –1 (low abundance) and 1 (high abundance). The yellow color indicates the more abundant patterns, while blue cells for less abundant RATC gene in that particular sample. The genes coding for MREP (multidrug resistance efflux pumps), CZCR (cobalt-zinc-cadmium resistance), BlaR (BlaR1 family regulatory sensor-transducer disambiguation); BLAC (beta-lactamase resistance), AR (arsenic resistance), RFL (resistance to fluoroquinolones), CH (copper homeostasis), CmeABC Operon (multidrug efflux pump in Campylobacter jejuni) had higher relative abundances than other RATC groups found in these CM samples. (B) The circular plot illustrates the diversity and relative abundance of the RATC genes detected among the microbiomes of the four different breeds (Local Zebu, LZ; Red Chattogram Cattle, RCC; Sahiwal, SW; Crossbred Holstein Friesian, XHF) of cows through SEED subsystems analysis. We found no significant correlation between the resistome and microbiome diversity in different breeds (P = 0.692). The association of the RATC genes according to breeds is shown by different colored ribbons and the relative abundances these genes are represented by inner blue colored bars. Part of the RATC functional groups are shared among microbes of the four breeds (XHF, LZ, SW, and RCC), and some are effectively undetected in the microbiomes of the other breeds. Abbreviations: CH, copper homeostasis; CHCT, copper homeostasis: copper tolerance; RCHC, resistance to chromium compounds; mdtABCD, the mdtABCD multidrug resistance cluster; OprN, mexe-mexf-oprn multidrug efflux system; MROP, mercury resistance to operon; MRS, methicillin resistance in Staphylococci; ZR, zinc resistance; BH, bile hydrolysis; ER, erythromycin resistance; ADCYS, adaptation to d-cysteine; SPVTL, Streptococcus pneumoniae vancomycin tolerance locus; STR, Streptothricin resistance; MAR Locus, multiple antibiotic resistance to locus; RVAN, resistance to vancomycin; MRD, mercuric reductase; LI, lysozyme inhibitors; AADNYL, aminoglycoside adenylyltransferases; mdtRP, multidrug resistance operon mdtRP of Bacillus; FR, Fosfomycin resistance; PSGCB, polymyxin synthetase gene cluster in Bacillus; OprM, mexA-mexB-oprm multidrug efflux system; CDR, cadmium resistance. Additional information is also available in Supplementary Material.
