FIGURE 5.
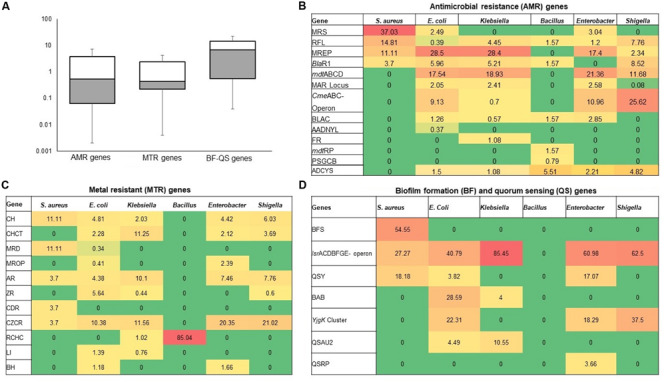
Heatmap comparison of antibiotics, metals, biofilm formation and quorum sensing genes found in the metagenome sequences (WMS) of six CM causing bacteria through SEED subsystems analysis in MG-RAST pipeline. (A) Diversity and relative abundance of the antimicrobial resistance (AMR), metal resistance (MTR), and biofilm formation (BF) and quorum sensing (QS) genes varied significantly (Kruskal–Wallis test, P = 0.029) among the study bacteria. (B) Relative abundance of AMR genes, (C) Relative abundance of MTR genes (D) Relative abundance of BF-QS genes. Values are colored in shades of green to yellow to red, indicating low (absent), medium and high abundance, respectively. Abbreviations: MRS, methicillin resistance in Staphylococci; RFL, resistance to fluoroquinolones; MREP, multidrug resistance to efflux pumps; BlaR, BlaR1 family regulatory sensor-transducer disambiguation; mdtABCD, the mdtABCD multidrug resistance cluster; MAR Locus, multiple antibiotic resistance; CmeABC Operon, Multidrug efflux pump in Campylobacter jejuni; BLAC, beta-lactamase resistance; AADNYL, aminoglycoside adenylyltransferases (Gentamycin resistance); FR, Fosfomycin resistance; mdtRP, multidrug resistance operon mdtRP of Bacillus; PSGCB, polymyxin synthetase gene cluster in Bacillus; BFS, biofilm formation in Staphylococcus, lsrACDBFGE operon, autoinducer 2 (AI-2) transport and processing; QSY, quorum sensing in Yersinia; BAB, biofilm adhesion biosynthesis; YjgK cluster, protein YjgK cluster linked to biofilm formation; QSAU2, quorum sensing: autoinducer-2 synthesis; QSRP, quorum sensing regulation in Pseudomonas; CH, copper homeostasis; CHCT, copper homeostasis: copper tolerance; MRD, mercuric reductase; MROP, mercury resistance to operon; AR, arsenic resistance; ZR, zinc resistance; CDR, cadmium resistance; CZCR, cobalt-zinc-cadmium resistance; ADCYS, adaptation to d-cysteine; RCHC, resistance to chromium compounds; LI, lysozyme inhibitors; BH, bile hydrolysis. More details about these genes can be found in the text and Supplementary Material.
