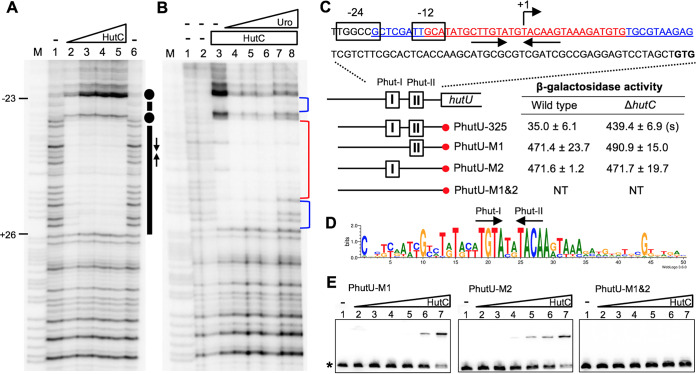
FIG 3.
Sequence determination of the HutC operator site in the PhutU promoter. (A) DNase I footprinting was performed using purified HutCHis6 and a 325-bp biotin-labeled probe, PhutU-325. Lane M, G+A marker; lanes 1 and 6, no HutCHis6; lanes 2 to 5, HutCHis6 added at 0.68, 2.39, 4.08, and 5.78 μM, respectively. The HutC-protected region and Phut half sites are indicated by bars and inverted arrows, respectively. Dots denote hypersensitive residues for DNase I cleavage. (B) Effects of urocanate on DNase I analysis of HutCHis6 (2.8 μM) and the PhutU-325 probe (2.0 μM). Lane M, G+A marker; lanes 1 and 2, no HutCHis6 and no urocanate; lanes 3 to 8, urocanate added at 0, 0.2, 0.5, 1, 2, and 3 mM, respectively. The strong and weak HutC-protected regions are marked with red and blue brackets, respectively. (C) The HutCHis6-protected region is underlined, and sequences of the strong and weak protected DNA regions are shown in red and blue, respectively. β-Galactosidase activity was measured in two genetic backgrounds (wild type versus ΔhutC) for lacZ fusion to the PhutU promoter and three derived mutant alleles lacking Phut-I and/or Phut-II repeats. “(s)” denotes a significant difference between the wild-type and ΔhutC strains as revealed by the Student t test (P < 0.01). NT, not tested. (D) Sequence logo generated from the alignment of PhutU and PhutF promoters from 40 representative Pseudomonas strains. (E) EMSA was performed using 325-bp biotin-labeled probes containing the wild-type or mutant alleles (Phut-I and/or Phut-II). Lanes 1 to 7, HutCHis6 added at 0, 37, 74, 148, 222, 296, and 555 nM, respectively.