FIGURE 3.
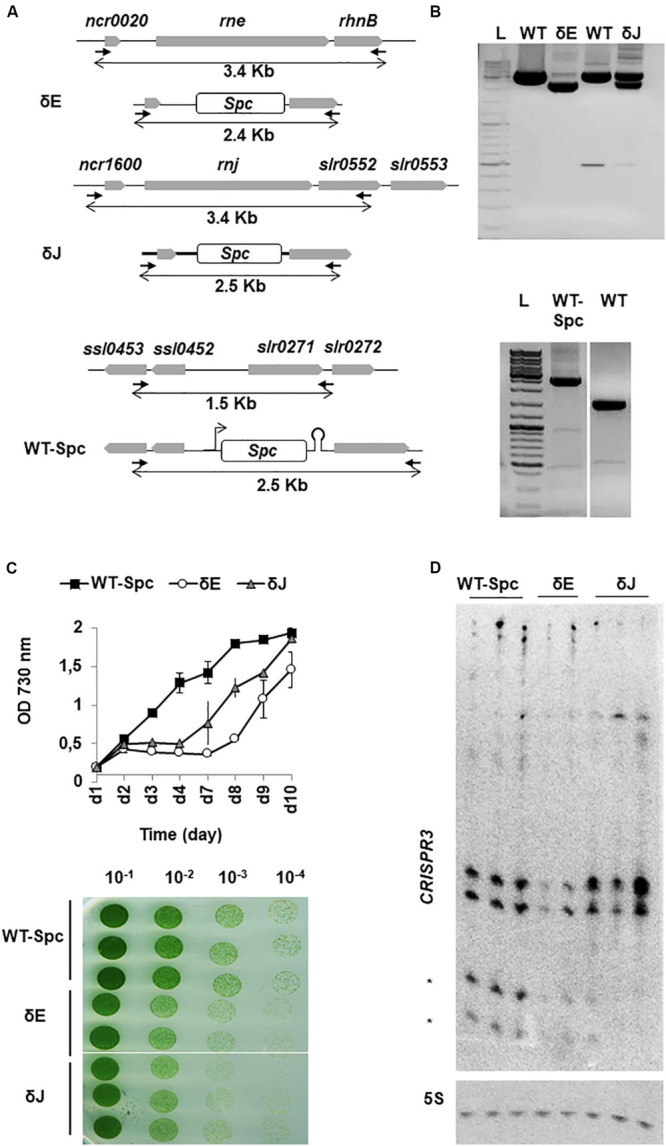
Deletion of rne and rnj genes in Synechocystis. (A) Schematic representation of mutant constructs. Genes are represented by gray boxes pointing in the direction of transcription. In δE and δJ strains the rne and rnj ORFs were replaced by a spc-resistance gene cassette (white box) by homologous recombination, while in the WT-Spc strain, the spc cassette was inserted between divergent transcriptional units. PCR primers used for segregation analysis are represented by black arrows; size of amplicons of WT and mutant chromosome loci are indicated by double arrows beneath each map. (B) PCR fragments of untransformed WT and mutant strains indicate the presence of the replaced (∼2.5 kb) and the WT (∼3.5 kb) chromosomal copies in δE and δJ strains (upper panel); the completely segregated insertion of the Spc cassette (∼2.5 kb) in the WT-Spc strain is shown in the lower panel. L = 1 kb DNA ladder. (C) WT-Spc, δE and δJ strains were grown in BG11 liquid and on solid medium supplemented with 200 μg/ml of Spc. Pre-cultures were diluted to OD730 nm = 0.2. Growth was monitored for 10 days with mean values ± SD calculated from two independent experiments. Cells suspended to OD730 nm = 0.2 were tenfold serially diluted in BG11 and spotted on solid medium. A representative of triplicate plates is shown. (D) Northern blot of CRISPR3 spacers 1–4. Mature crRNAs are indicated by asterisks. 5S rRNA (5S) was used as a loading control.
