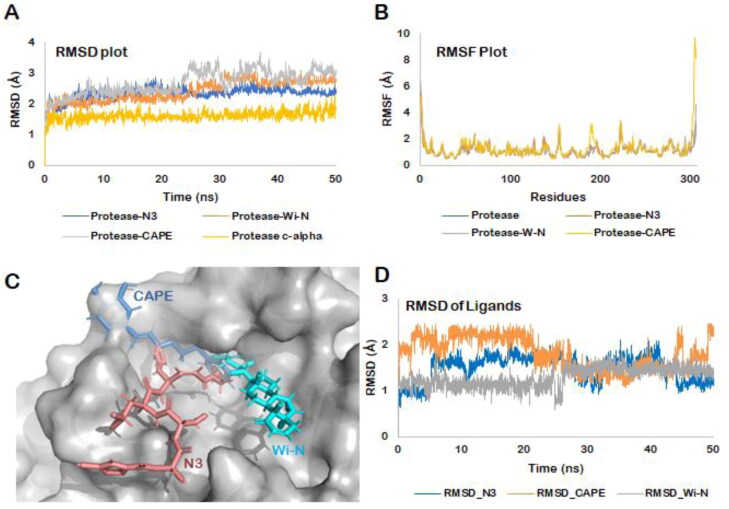
Figure 4.
(A) RMSD of the protein backbone along the simulation trajectory for the protein alone and all the docked complexes. The overall structure of Mpro did not change much after the binding of Wi-N or CAPE when compared to N3 inhibitor. (B) RMSF of the amino acids comprising the Mpro. No abrupt fluctuations were observed in any region of the protein with or without the three ligands. (C) Superimposition of the three docked complexes. All the three small molecules- N3 protease inhibitor, Wi-N and CAPE were bound in the same site suggesting their similar mechanism of action. (D) RMSD plot for all the three ligands over the entire simulation trajectory. Similar to N3 inhibitor, Wi-N and CAPE stayed bound in almost the same docked pose throughout the simulation run.