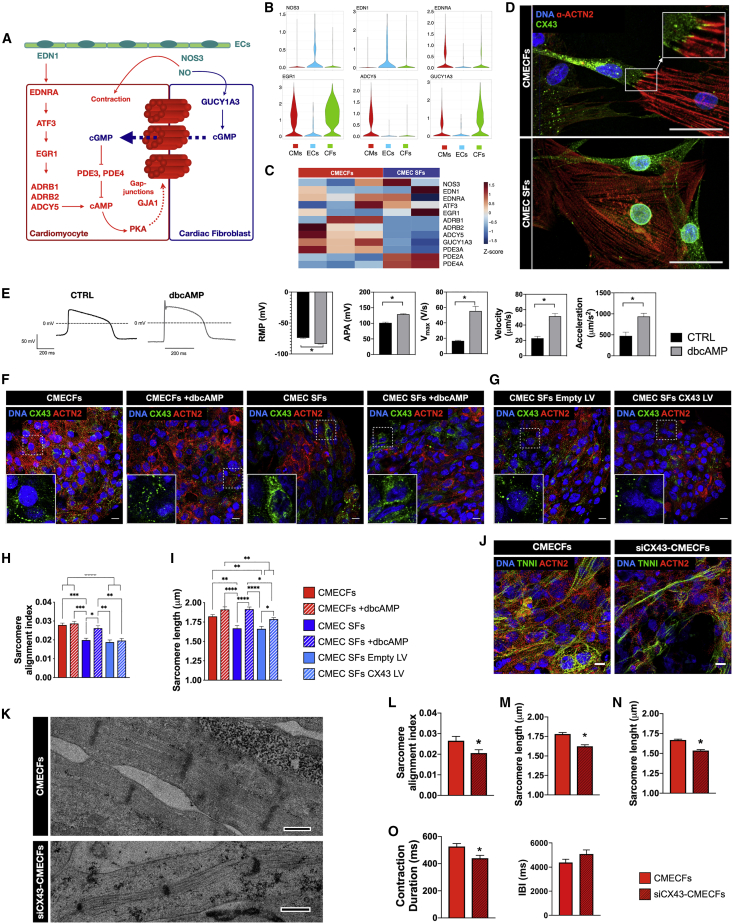
Figure 6.
Mechanisms Underlying hiPSC-CM Maturation in Microtissues with CFs and ECs
(A) Proposed mechanism underlying hiPSC-CM maturation in MTs with CFs and ECs: ECs (green) secrete EDN1 that activates β-adrenergic signaling and adenylyl cyclase in CMs (red), increasing intracellular cAMP levels, which can enhance CX43 gap junction formation. ECs also secrete NO that activates cGMP pathway in CFs (blue). cGMP can shuttle to CMs via gap junctions (dotted blue arrow), sustaining cAMP in CMs.
(B) Violin plots showing (log-transformed) expression of NOS3, EDN1, EDNRA, EGR1, ADCY5, and GUCY1A3 in CMECFs based on scRNA-seq.
(C) Heatmap showing selected gene expression from bulk RNA-seq of CMECFs and CMEC SFs.
(D) Immunofluorescence analysis of CX43 (green) and ACTN2 (red) in hiPSC-CMs and fibroblasts dissociated from CMECFs and CMEC SFs MTs. White arrows indicate coupling between hiPSC-CFs and hiPSC-CMs. SFs do not couple with hiPSC-CMs. Nuclei are stained with DAPI (blue). Scale bar: 50 μm.
(E) Representative AP traces of untreated (CTRL, black) and 72-h-dbcAMP-treated (dbcAMP, gray) CTRL1 hiPSC-CMs, with quantification of RMP, APA, Vmax, contraction velocity, and acceleration. n > 10; dissociated cells per group; ∗p < 0.001. Data are mean ± SEM. Student’s t test is shown.
(F and G) Representative immunofluorescence images of CX43 (green) and ACTN2 (red) in MTs from CTRL1 hiPSCs, either untreated (CMECFs, CMEC SFs) or treated for 7 days with dbcAMP (CMECFs +dbcAMP, CMEC SFs +dbcAMP; F) and MTs from CTRL1 hiPSC containing either SFs transduced with control lentivirus (LV) (CMEC SFs empty LV) or lentivirus containing CX43 LV (CMEC SFs CX43 LV; G). Nuclei are stained with DAPI (blue). Scale bar: 10 μm. Insets are magnifications of framed areas to show CX43 distribution.
(H and I) Sarcomere organization (sarcomere alignment index; H) and sarcomere length (I) from immunofluorescence analysis of MTs from CTRL1 hiPSCs. n = 30; areas from 3 MTs per group; ∗∗p < 0.05; ∗∗∗p < 0.005; ∗∗∗∗p < 0.0001. Data are mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test is shown.
(J) Representative immunofluorescence images of cardiac sarcomeric proteins ACTN2 (red) and TNNI (green) in CMECFs generated from CTRL1 CMECFs containing either untreated hiPSC-CFs (CMECFs) or hiPSC-CFs transduced with CX43-shRNA (siCX43-CMECFs). Nuclei are stained with DAPI (blue). Scale bar: 10 μm.
(K) TEM showing sarcomeres in CMECFs and siCX43-CMECFs. Scale bar: 0.5 μm.
(L–N) Quantification of sarcomere organization (sarcomere alignment index; L) and sarcomere length (M) from immunofluorescence analysis in MTs (n = 60; areas from 4 MTs per group; ∗p < 0.05) and of sarcomere length in MTs from TEM (-n; n > 117; areas from 3 independent stitches per group; ∗p < 0.0001). Data are shown as mean ± SEM. Student’s t test is shown.
(O) Contraction duration (left) and IBI (right) measured in spontaneously beating MTs. n > 40; MTs from 3 independent batches per group; ∗p < 0.05. Student’s t test is shown. Data are shown as mean ± SEM.