Abstract
Osteoarthritis (OA) is a common chronic joint disease affected by environmental and genetic factors. The LTBP3 gene may be involved in the occurrence and development of OA by regulating TGF-β activity and the TGF-β signaling pathway. A total of 2780 study subjects, including 884 hip OA cases and 1896 controls, were recruited. Nine tag single-nucleotide polymorphisms (SNPs) located within the LTBP3 gene region were selected for genotyping. Genetic association analyses were performed at both the genotypic and allelic levels. GTEx data were extracted to investigate the functional consequence of significant SNPs. SNP rs10896015 was significantly associated with the risk of hip OA at both the genotypic (P=0.0019) and allelic levels (P=0.0009). The A allele of this SNP was significantly associated with a decreased risk of HOA (OR [95%CI] = 0.79 [0.69–0.91]). This SNP was also significantly associated with the clinical severity of hip OA. SNP rs10896015 could affect the gene expression of 11 genes, including LTBP3, in multiple human tissues based on GTEx data. We obtained evidence for a genetic association between the LTBP3 gene and hip OA susceptibility and clinical severity based on Chinese Han populations. Our findings replicated the association signals reported by a recent genome-wide association study and deepen the basic understanding of osteoarthritis pathology.
Keywords: genetic association, hip osteoarthritis, LTBP3 gene, single nucleotide polymorphism
Introduction
Osteoarthritis (OA) is a common chronic joint disease characterized by primary or secondary degeneration of articular cartilage and bone hyperplasia [1]. Of these, articular cartilage degeneration is the earliest and most important pathological change [2]. The most common pathological changes of OA occur in the knee joint and hip joint. Although the prevalence of hip OA (HOA) is lower than knee OA, HOA is more likely to cause disability and death in the elderly [3]. The lifetime risk of developing HOA is approximately 25%, which leads to a socioeconomic and healthcare burden [4]. Similar to other chronic and complex diseases, HOA is also affected by environmental and genetic factors. Age and gender can influence the incidence of HOA, and multiple genetic studies have confirmed the importance of genetic factors [5]. Twin and family studies indicated that the heritability for HOA is approximately 60% [6]. Hence, it is necessary to identify the genes responsible for susceptibility to HOA through candidate gene studies.
Recently, single-nucleotide polymorphism (SNP) rs10896015 in the latent TGF-β binding protein3 (LTBP3) gene was significantly associated with HOA in the British population through a genome-wide association study (GWAS) that included 77,052 OA patients and 378,169 controls [7]. To the best of our knowledge, this is the first association study between the HOA and LTBP3 genes. LTBP3 is a member of the latent TGF-β binding protein, which has four isoforms (LTBP1–4) in the human genome [8]. The cysteine-rich domain of LTBP-1, 3, and 4 can covalently bind latent TGF-β [9]. Through the use of human osteoarthritic osteoblasts, researchers have found increased levels of TGF-β, which may up-regulate cartilage matrix-degrading enzyme expression and ultimately result in OA [10,11]. In addition, researchers have also found increased expression levels of TGF-β in articular cartilage and osteophytes in an OA mouse model, and this phenomenon can be reversed by endogenous TGF-β [12]. These results suggested that expression changes in TGF-β could affect OA directly. A recent study demonstrated that up-regulated LTBP1 caused activation of the TGF-β signaling pathway in human OA fibroblast-like synoviocytes, which may lead to the aggravation of OA [13]. Hence, the LTBP3 gene may also be involved in the occurrence and development of OA by regulating TGF–β activity and the TGF-β signaling pathway. However, few studies have focused on the relationship between the LTBP3 gene and HOA, and the only association study was conducted in individuals of European ancestry. To confirm whether the common variants in the LTBP3 gene were universally associated with the risk of HOA, we systematically investigated the association of the LTPB3 gene with the risk of HOA in the Han Chinese population in the present study.
Methods
Study subjects
A total of 2780 study subjects, including 884 HOA cases and 1896 controls, were recruited from the Second Hospital of Xi'an Jiaotong University from March 2014 to February 2019. HOA patients were diagnosed by three independent physicians based on criteria published by the American College of Rheumatology. Demographic information, including age, gender, BMI, smoking and alcohol consumption, was collected through a questionnaire. HOA patients with Kellgren–Lawrence (KL) grading scores of 2 or more were included. Study subjects with no symptoms and family history of arthritis or any other joint-related disorders were enrolled as controls. Study subjects were excluded if they had (1) inflammatory arthritis, (2) post-traumatic or post-septic arthritis, (3) skeletal or developmental dysplasia, or (4) systemic or organic diseases. The present study was approved by the institutional review board of the Second Hospital of Xi’an Jiaotong University. Informed consent forms were signed and obtained from all study subjects.
SNP selection and genotyping
For SNP selection, 1000 Genomes Chinese Han Beijing (CHB) data were used as reference data. We extracted all SNPs with MAFs greater than 0.03 within the LTBP3 gene region in 1000 Genome CHB data, resulting in a SNP set of 19. Then, tag SNPs (r2≥0.8) were identified within this SNP set based on the method proposed by Gabriel et al. [14]. Finally, a total of 9 tag SNPs located within the LTBP3 gene region were selected for genotyping (Supplementary Table S1). We extracted genomic DNA from peripheral blood leukocytes according to the manufacturer’s protocol (Genomic DNA Kit, Axygen Scientific, Inc., CA, U.S.A.). A high-throughput Sequenom MassARRAY platform (Sequenom, San Diego, CA, U.S.A.) was utilized for SNP genotyping. Briefly, the signals from the platform were automatically analyzed using Sequenom Typer 4.0 software, and genotype data were generated from the processed results [15]. To estimate the genotyping quality, 5% of random samples were repeated for genotyping. With a concordance rate of 100%, the quality of genotyping data was confirmed. The case/control status of the samples was blinded to the technicians during the genotyping process [16].
Statistical analyses
We have conducted a comprehensive power statistical analysis for the present study using GAS power calculator (http://csg.sph.umich.edu/abecasis/cats/gas_power_calculator/). The results indicated that our sample size could achieve approximately 71% statistical power for genetic association (Supplementary Table S2 and Supplementary Figure S1). Single marker-based association analyses were conducted at both allelic and genotypic levels. χ2 tests were performed for both levels of analyses. Further association analysis was conducted between significant SNPs and KL grading scores in the HOA patient group. Genetic association analyses were performed by Plink [17]. Linkage disequilibrium structures of our selected SNPs were visualized using Haploview [18]. Bonferroni corrections were applied to address multiple comparisons. In general, the threshold of the P-value for single marker-based association analyses was 0.05/9≈0.006. To further investigate the functional consequences of the significant hits identified from genetic association analyses, we extracted the expression quantitative trait loci (eQTL) data obtained from multiple human tissues from the GTEx database [19].
Results
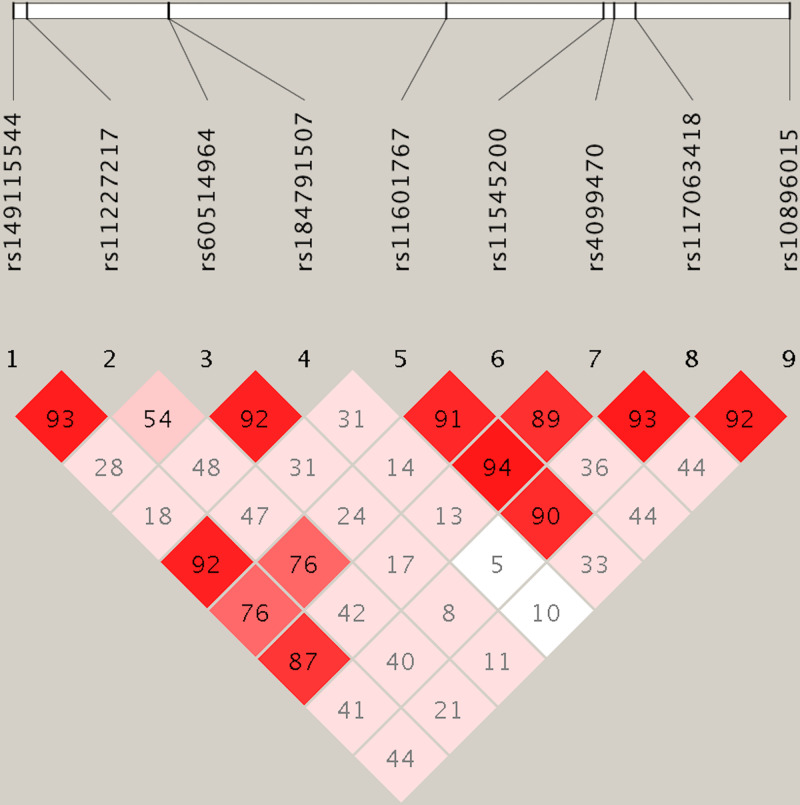
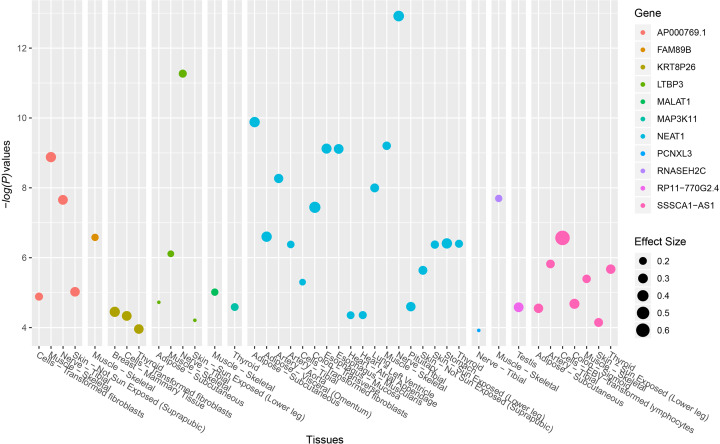
A total of 884 HOA cases and 1896 healthy controls were recruited in the present study (Table 1). No significant differences were identified between HOA patients and healthy controls in multiple demographic and clinical variables, including age, gender, BMI, smoking and alcohol consumption status. In the 884 HOA patients, there were 372 with KL scores of 2 (42%), 325 with KL scores of 3 (37%) and 187 with KL scores of 4 (21%). SNP rs10896015, an intronic SNP located in gene LTBP3, was significantly associated with the disease status of HOA at both the genotypic (P=0.0019) and allelic levels (P=0.0009). The A allele of this SNP, which was the minor allele, was significantly associated with a decreased risk of HOA (OR [95%CI] = 0.79 [0.69–0.91], Table 2). We further examined the association between SNP rs10896015 and KL grading scores in the HOA patients. We identified that this SNP was significantly associated with KL grading score (Table 3). There were significantly more patients with GG genotypes in the KL-4 group than in the KL-2 and KL-3 groups (69% vs. 62% vs. 62%, respectively). The LD structure of the nine selected SNPs is shown in Figure 1. We explored SNP rs10896015 in the GTEx database and identified that the genotypes of this SNP were significantly associated with the gene expression of LTBP3 in various human tissues (Supplementary Table S3 and Figure 2). Interestingly, in addition to LTBP3, SNP rs10896015 was also significantly associated with the gene expression levels of several other genes, including AP000769.1, FAM89B, KRT8P26, MALAT1, MAP3K11, NEAT1, PCNXL3, RNASEH2C, RP11-770G2.4, and SSSCA1-AS1.
Table 1. Demographic information of the study subjects.
| Variables | Cases (N=884) | Controls (N=1896) | Statistics | P-value |
|---|---|---|---|---|
| Age, years | 61.8 ± 7.9 | 61.7 ± 8.2 | T = 0.35 | 0.73 |
| BMI, kg/m2 | 25.9 ± 1.4 | 25.8 ± 1.6 | T = 1.56 | 0.12 |
| Gender (%) | ||||
| Male | 387 (44) | 826 (44) | ||
| Female | 497 (56) | 1070 (56) | χ2 = 0.0041 | 0.95 |
| Smoking (%) | ||||
| Yes | 262 (30) | 559 (29) | ||
| No | 622 (70) | 1337 (71) | χ2 = 0.0015 | 0.97 |
| Alcohol consumption (%) | ||||
| Yes | 229 (26) | 488 (26) | ||
| No | 655 (74) | 1408 (74) | χ2 = 0.0022 | 0.96 |
| KL grade (%) | ||||
| KL-2 | 372 (42) | |||
| KL-3 | 325 (37) | |||
| KL-4 | 187 (21) |
Table 2. Results of single marker-based association analyses.
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | G | A | ||||||||
| rs149115544 | 65539637 | Cases | 6 | 104 | 774 | 116 | 1652 | |||||
| Controls | 9 | 220 | 1667 | 0.49 | 0.78 | 238 | 3554 | 0.16 | 0.69 | 1.05[0.83–1.32] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | T | C | ||||||||
| rs11227217 | 65539931 | Cases | 27 | 267 | 590 | 321 | 1447 | |||||
| Controls | 65 | 576 | 1255 | 0.29 | 0.87 | 706 | 3086 | 0.17 | 0.68 | 0.97[084–1.12] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | G | A | ||||||||
| rs60514964 | 65542976 | Cases | 9 | 129 | 746 | 147 | 1621 | |||||
| Controls | 15 | 262 | 1619 | 0.69 | 0.71 | 292 | 3500 | 0.63 | 0.43 | 1.09[0.88–1.34] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | ||||||||
| rs184791507 | 65542980 | Cases | 4 | 78 | 802 | 86 | 1682 | |||||
| Controls | 6 | 163 | 1727 | – | 0.80 | 175 | 3617 | 0.17 | 0.68 | 1.06[0.81–1.38] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||||||||
| rs11601767 | 65548911 | Cases | 115 | 407 | 362 | 637 | 1131 | |||||
| Controls | 254 | 884 | 785 | 0.25 | 0.88 | 1392 | 2400 | 0.24 | 0.62 | 0.97[0.86–1.09] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | ||||||||
| rs11545200 | 65552280 | Cases | 5 | 95 | 784 | 105 | 1663 | |||||
| Controls | 6 | 201 | 1689 | 0.97 | 0.62 | 213 | 3579 | 0.23 | 0.63 | 1.06[0.83–1.35] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | T | C | ||||||||
| rs4099470 | 65552515 | Cases | 11 | 196 | 677 | 218 | 1550 | |||||
| Controls | 28 | 425 | 1443 | 0.27 | 0.88 | 481 | 3311 | 0.14 | 0.71 | 0.97[0.82–1.15] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | ||||||||
| rs117063418 | 65552969 | Cases | 5 | 94 | 785 | 104 | 1664 | |||||
| Controls | 8 | 197 | 1,691 | 0.31 | 0.86 | 213 | 3579 | 0.16 | 0.69 | 1.05[0.82–1.34] | ||
| SNP | Position | Status | Genotypes | *χ2 | P-value | Alleles | χ2 | P-value | OR [95%CI] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | ||||||||
| rs10896015 | 65556254 | Cases | 30 | 292 | 562 | 352 | 1416 | |||||
| Controls | 116 | 675 | 1105 | 12.48 | 0.0019 | 907 | 2885 | 11.07 | 0.0009 | 0.79[0.69–0.91] | ||
The threshold of the P-value is 0.05/9≈0.006. Significant markers are highlighted in bold.
Fisher’s exact test was applied for sparse contingency tables.
Table 3. Results of association between KL grading scores and genotypes of SNP rs10896015.
| KL grade | Genotypes | χ2 | P-value | ||
|---|---|---|---|---|---|
| AA | AG | GG | |||
| KL-2 (%) | 9 (3) | 130 (35) | 233 (62) | ||
| KL-3 (%) | 7 (2) | 118 (36) | 200 (62) | ||
| KL-4 (%) | 14 (7) | 44 (24) | 129 (69) | 19.45 | 0.0006 |
Figure 1. Linkage disequilibrium plot of the selected SNPs.
Values of D’ are indicated in each cell.
Figure 2. eQTL signals of SNP rs10896015 achieved genome-wide significance.
Data were extracted from the GTEx database.
Discussion
In the present study, we found that an intronic SNP of the LTBP3 gene, SNP rs10896015, was significantly associated with HOA disease status based on study subjects with Chinese Han ancestry. To the best of our knowledge, our study was the first to link LTBP3 to HOA in a Chinese Han population. SNP rs10896015 was significantly associated with HOA in a recently published GWAS using UK Biobank data, which are mainly based on European populations [7]. The effect direction of SNP rs10896015 was the same in our study and the recent GWAS. Thus, our study could be considered as a validation for this recent GWAS. In addition, in the present study, we took it one step further to show that SNP rs10896015 was not only associated with the disease status of HOA but also significantly associated with the clinical severity of HOA.
The product of LTBP3 is a member of latent TGF-β binding proteins. A large number of studies have indicated that TGF-β plays an important role in the formation of articular chondrocytes and the homeostasis of cartilage matrix [20,21]. Cardiovascular defects were found in an LTBP3 knockdown zebrafish model, which had been rescued by a constitutively active TGFβ type 1 receptor [22]. This result suggests that LTBP3 may also be a regulator of TGF-β. Moreover, LTBP3 (LTBP3-/-) knockout mice showed the same pathological changes in joints as OA, such as osteophytes, ossified and fibrotic areas on the articular surface and the absence of articular cartilage [23]. The same result was consistent with the perturbed TGF-β signaling pathway in mouse long bones, which further strengthens the evidence linking LTBP3 and TGF-β [24].
With the fast development of target sequencing, numerous susceptibility variants of complex diseases have been identified, such as schizophrenia [25–27] (Zhang and others 2018; Han and others 2019; Guan and others 2020a). Given that it is difficult to draw reliable conclusions only from single SNP association analyses [28–32] (Zhu and others 2016; Sun and others 2017; Zhang and others 2017; Li and others 2018; Guan and others 2020b), although SNP rs10896015 is an intronic SNP that cannot alter the structure of the protein encoded by LTBP3, our analyses using GTEx data confirmed that it was significantly associated with the gene expression of LTBP3 in multiple human tissues. Therefore, SNP rs10896015 might be a DNA variant with functional significance but not just a surrogate of some underlying DNA variants that were not genotyped in this present study. In addition, the eQTL analysis using GTEx data also provides interesting results. It seemed that SNP rs10896015 was an eQTL for LTBP3 and for several other genes located near SNP rs10896015 and the LTBP3 gene. Of these, some genes, such as AP000769.1 and RP11-770G2.4, had unknown functions (most likely some nonfunctional peptides) and were not worthy of discussion. However, a long noncoding gene, NEAT1, was of particular interest. First, SNP rs10896015 was a significant eQTL signal in a large number of human tissues for NEAT1. Second, NEAT1 was linked to osteogenic differentiation in human bone marrow-derived mesenchymal stem cells in a recent study and therefore might play a role in the metabolism of bone tissues [33]. In this sense, we have now identified a significant SNP for HOA; however, the genetic location of this SNP was unclear. One strategy was to physically map this SNP to the LTBP3 gene because this SNP is located within its gene region. This is a common SNP-gene mapping strategy used in most (if not all) GWAS. On the other hand, this SNP could probably be functionally mapped to other genes, such as NEAT1, when eQTL data were integrated and considered. In the present study, our data could not provide enough information to address this issue. In the future, experimental research based on animal models will be conducted to clarify this point.
Our study has from several limitations. First, as a genetic association study, the association signals identified in our study might be confounded by population stratification. Unfortunately, as a candidate gene-based association study, we could use common methods utilized in GWAS, such as principal component analysis, to control this most commonly identified confounder in population genetics studies. Another limitation is that the information coverage of our selected SNPs might be insufficient to explore the whole functional regions of LTBP3. Only 9 tag SNPs within the gene region were genotyped in this present study, and SNPs located at ±20 kb up/downstream of LTBP3, which are important functional regions for the regulation of gene expression, were not investigated in this study. This strategy of SNP selection enabled us to maximize the information coverage under limited experimental expense, although it might miss significant functional regions of the targeted gene.
In summary, in the present study, we obtained evidence for a genetic association between LTBP3 and HOA disease status and clinical severity based on Chinese Han populations. Our findings replicated the association signals reported by recent GWAS and could deepen the basic understanding of osteoarthritis pathology.
Supplementary Material
Abbreviations
- eQTL
expression quantitative trait loci
- HOA
hip OA
- KL
Kellgren–Lawrence
- OA
osteoarthritis
- SNP
single-nucleotide polymorphism
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
Tianyun Zhao and Jibin Liu conceived and designed the study. Tianyun Zhao carried out candidate SNPs selection and statistical analyses. Tianyun Zhao, Junji Zhao, Chi Ma and Jie Wei conducted subject screening. Tianyun Zhao, Junji Zhao and Bo Wei contributed to the collection and preparation of DNA samples. Tianyun Zhao wrote the paper.
References
- 1.Kijowski R., Blankenbaker D., Stanton P. et al. (2006) Correlation between radiographic findings of osteoarthritis and arthroscopic findings of articular cartilage degeneration within the patellofemoral joint. Skeletal Radiol. 35, 895–902 10.1007/s00256-006-0111-7 [DOI] [PubMed] [Google Scholar]
- 2.Chen D., Shen J., Zhao W. et al. (2017) Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 5, 16044 10.1038/boneres.2016.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aresti N., Kassam J., Nicholas N. et al. (2016) Hip osteoarthritis. BMJ 354, i3405 10.1136/bmj.i3405 [DOI] [PubMed] [Google Scholar]
- 4.Culliford D.J., Maskell J., Kiran A. et al. (2012) The lifetime risk of total hip and knee arthroplasty: results from the UK general practice research database. Osteoarthritis Cartilage 20, 519–524 10.1016/j.joca.2012.02.636 [DOI] [PubMed] [Google Scholar]
- 5.Spector T.D. and MacGregor A.J. (2004) Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage 12, S39–S44 10.1016/j.joca.2003.09.005 [DOI] [PubMed] [Google Scholar]
- 6.MacGregor A.J., Antoniades L., Matson M. et al. (2000) The genetic contribution to radiographic hip osteoarthritis in women: results of a classic twin study. Arthritis Rheum. 43, 2410–2416 [DOI] [PubMed] [Google Scholar]
- 7.Tachmazidou I., Hatzikotoulas K., Southam L. et al. (2019) Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat. Genet. 51, 230–236 10.1038/s41588-018-0327-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handford P.A., Downing A.K., Reinhardt D.P. et al. (2000) Fibrillin: from domain structure to supramolecular assembly. Matrix Biol.: J. Int. Soc. Matrix Biol. 19, 457–470 10.1016/S0945-053X(00)00100-1 [DOI] [PubMed] [Google Scholar]
- 9.Koli K., Saharinen J., Hyytiainen M. et al. (2001) Latency, activation, and binding proteins of TGF-beta. Microsc. Res. Tech. 52, 354–362 [DOI] [PubMed] [Google Scholar]
- 10.Massicotte F., Lajeunesse D., Benderdour M. et al. (2002) Can altered production of interleukin-1β, interleukin-6, transforming growth factor-β and prostaglandin E 2 by isolated human subchondral osteoblasts identify two subgroups of osteoarthritic patients. Osteoarthritis Cartilage 10, 491–500 [DOI] [PubMed] [Google Scholar]
- 11.Moldovan F., Pelletier J.P., Hambor J. et al. (2014) Collagenase-3 (matrix metalloprotease 13) is preferentially localized in the deep layer of human arthritic cartilage in situ: in vitro mimicking effect by transforming growth factor beta. Arthritis Rheum. 40, 1653–1661 10.1002/art.1780400915 [DOI] [PubMed] [Google Scholar]
- 12.Scharstuhl A., Glansbeek H.L., van Beuningen H.M. et al. (2002) Inhibition of endogenous TGF-beta during experimental osteoarthritis prevents osteophyte formation and impairs cartilage repair. J. Immunol. 169, 507–514 10.4049/jimmunol.169.1.507 [DOI] [PubMed] [Google Scholar]
- 13.Wang X.L., Dong C., Li N. et al. (2018) Modulation of TGF- activity by latent TGF–binding protein 1 in human osteoarthritis fibroblast-like synoviocytes. Mol. Med. Rep. 17, 1893–1900 [DOI] [PubMed] [Google Scholar]
- 14.Gabriel S.B., Schaffner S.F., Nguyen H. et al. (2002) The structure of haplotype blocks in the human genome. Science 296, 2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- 15.Guan F., Zhang C., Wei S. et al. (2012) Association of PDE4B polymorphisms and schizophrenia in Northwestern Han Chinese. Hum. Genet. 131, 1047–1056 10.1007/s00439-011-1120-8 [DOI] [PubMed] [Google Scholar]
- 16.Guan F., Zhang B., Yan T. et al. (2014) MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr. Res. 152, 97–104 10.1016/j.schres.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 17.Purcell S., Neale B., Todd-Brown K. et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett J.C., Fry B., Maller J. et al. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 19.Lonsdale J., Thomas J., Salvatore M. et al. (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C.C., Silverman R.M., Shen J. et al. (2018) Distinct metabolic programs induced by TGF-beta 1 and BMP2 in human articular chondrocytes with osteoarthritis. J. Orthop. Transl. 12, 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhen G.H. and Cao X. (2014) Targeting TGF beta signaling in subchondral bone and articular cartilage homeostasis. Trends Pharmacol. Sci. 35, 227–236 10.1016/j.tips.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yong Z., Cashman T.J., Nevis K.R. et al. (2012) Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dabovic B., Chen Y., Colarossi C. et al. (2002) Bone abnormalities in latent TGF-beta binding protein (Ltbp)-3-null mice indicate a role for Ltbp-3 in modulating TGF-beta bioavailability. J. Cell Biol. 156, 227–232 10.1083/jcb.200111080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jie S., Jia L., Baoli W. et al. (2013) Deletion of the Transforming Growth Factor β Receptor Type II Gene in Articular Chondrocytes Leads to a Progressive Osteoarthritis-like Phenotype in Mice. Arthritis Rheumatism 65, 3107–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T.X., Zhu L., Ni T. et al. (2018) Voltage-gated calcium channel activity and complex related genes and schizophrenia: A systematic investigation based on Han Chinese population. J. Psychiatr. Res. 106, 99–105 10.1016/j.jpsychires.2018.09.020 [DOI] [PubMed] [Google Scholar]
- 26.Han W., Zhang T.X., Ni T. et al. (2019) Relationship of common variants in CHRNA5 with early-onset schizophrenia and executive function. Schizophr. Res. 206, 407–412 10.1016/j.schres.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 27.Guan F., Ni T., Han W. et al. (2020) Evaluation of the relationships of the WBP1L gene with schizophrenia and the general psychopathology scale based on a case–control study. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 183, 164–171 10.1002/ajmg.b.32773 [DOI] [PubMed] [Google Scholar]
- 28.Zhu L., Li J., Dong N. et al. (2016) mRNA Changes in Nucleus Accumbens Related to Methamphetamine Addiction in Mice. Sci. Rep. 6, 36993 10.1038/srep36993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun H., Luo C., Chen X. and Tao L. (2017) Assessment of Cognitive Dysfunction in Traumatic Brain Injury Patients: A Review. Forensic Sci. Res. 2, 174–179 10.1080/20961790.2017.1390836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Gong Q., Feng X., Zhang D. and Quan L. (2017) Astrocytic Clasmatodendrosis in the Cerebral Cortex of Methamphetamine Abusers. Forensic Sci. Res. 2, 139–144 10.1080/20961790.2017.1280890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Zhu L., Guan F. et al. (2018) Relationship Between Schizophrenia and Changes in the Expression of the Long Non-Coding RNAs Meg3, Miat, Neat1 and Neat2. J. Psychiatr. Res. 106, 22–30 10.1016/j.jpsychires.2018.09.005 [DOI] [PubMed] [Google Scholar]
- 32.Guan F., Zhang T., Han W. et al. (2020) Relationship of SNAP25 Variants With Schizophrenia and Antipsychotic-Induced Weight Change in Large-Scale Schizophrenia Patients. Schizophr. Res. 215, 250–255 10.1016/j.schres.2019.09.015 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y.Z., Chen B., Li D.Z. et al. (2019) LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic differentiation in human bone marrow-derived mesenchymal stem cells. Pathol. Res. Pract. 215, 525–531 10.1016/j.prp.2018.12.034 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.