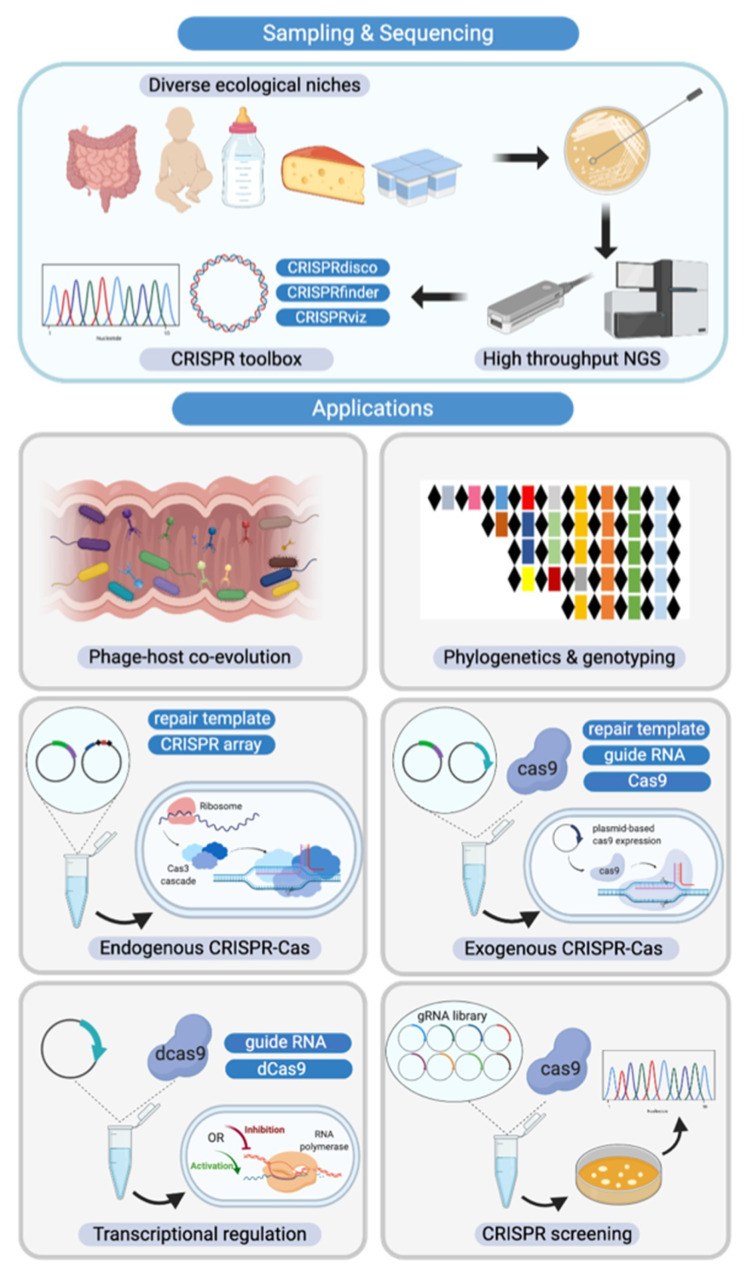
Figure 7.
Diverse CRISPR-Cas system applications in Bifidobacterium. (Top Panel) Novel Bifidobacterium species and CRISPR-Cas systems discovered through high-throughput next generation sequencing, assisted by a plethora of bioinformatic analysis tools; (Bottom Panel) From left to right and then top to bottom. CRISPR-Cas systems serve as the bacterial adaptive immune system, influencing the predator-prey dynamics between phages and commensals such as Bifidobacterium in the human gastrointestinal tract. CRISPR arrays can also be used as unique genetic markers for strain identification and genotyping. As discussed in this study, CRISPR-Cas systems are highly abundant in bifidobacterial genomes. With in-depth analysis of CRISPR elements such as crRNA, tracrRNA, and PAM reported here, we have built the platform for repurposing these endogenous CRISPR-Cas systems as genome editing tools. In the case of absence of CRISPR-Cas systems or lack of functionality, exogenous CRISPR-Cas systems can be delivered using plasmid-based systems. Beyond genome editing, alternative CRISPR-Cas systems with deactivated nucleases can serve as transcriptional regulation tools. In light of next generation sequencing (NGS), guide RNA libraries can be prepared to screen natural Bifidobacterium variants in a high-throughput fashion, bypassing the genetic modification route that can require strict regulation.