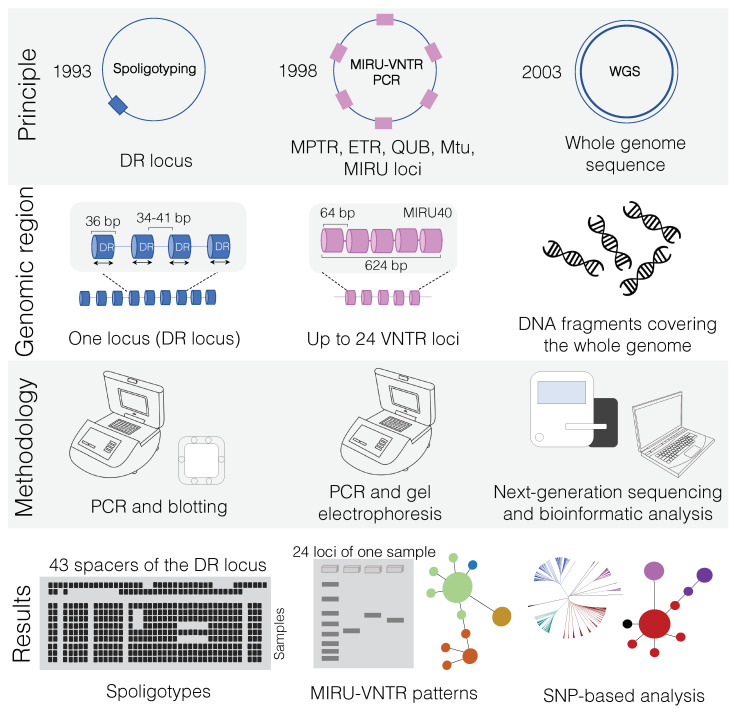
Figure 2.
Overview of main genotyping techniques (Spoligotyping and MIRU-VNTR) and whole-genome sequencing (WGS) used for transmission cluster investigation of Mycobacterium bovis. In “principle”, squares denote the quantity of specific genetic markers (i.e., DR locus and VNTR) on M. bovis genomes. While spoligotyping is based on a unique locus, MIRU-VNTR PCR amplifies genetic targets from multiple regions of the genome (up to 24 loci). In contrast, WGS uses information from the whole-genome sequence. Dates refer to the year in which each technique was developed. In “genomic region”, the MIRU40 locus is shown as an example of one of the 24 loci that can be used in MIRU-VNTR PCR. In “results”, the spoligotyping membrane is depicted accommodating several samples simultaneously, owing to the high-throughput capability of this technique (up to 45 samples can be simultaneously analyzed). In MIRU-VNTR PCR, although many samples can be amplified at once, each sample can occupy up to 24 wells in an agarose gel, so many electrophoresis runs may be needed depending on the laboratory. MIRU-VNTR databases can subsequently be used to generate a minimum spanning tree. WGS is a high-throughput technique that will lead to single nucleotide polymorphism (SNP)-based analysis. The same generated data can also be used to detect spoligotype and MIRU-VNTR patterns (see text). WGS results can be used to evaluate transmission clusters as well as phylogenetic relationships among the sequenced genomes. WGS: whole-genome sequencing; MIRU-VNTR: mycobacterial interspersed repetitive unit-variable-number tandem repeat typing; PCR: polymerase chain reaction; DR: direct repeat.