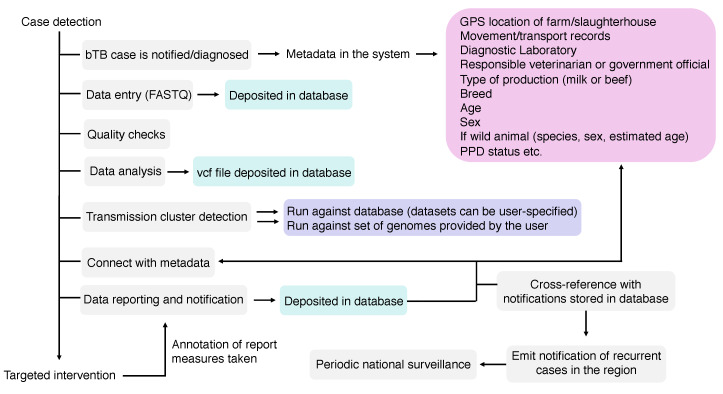
Figure 5.
Components of a Mycobacterium bovis whole-genome sequencing (WGS) pipeline. In grey: components of the pipeline; in green: files that will compose the sequence and analysis databases (raw reads—FASTQ—and vcf file); in pink: metadata, which can also compose a metadata database linked to each FASTQ and vcf file; in purple: the possibilities of genome comparisons: (i) a user can choose to compare the genome against all genomes of the database or against a subset of genomes composing the database; or (ii) a user can input several genomes that can be compared against each other or with other genomes deposited in the database. An ideal pipeline would also allow periodic national surveillance reports, emitting alerts of newly detected clusters or outbreaks in certain regions that warrant further attention according to user-specified thresholds.