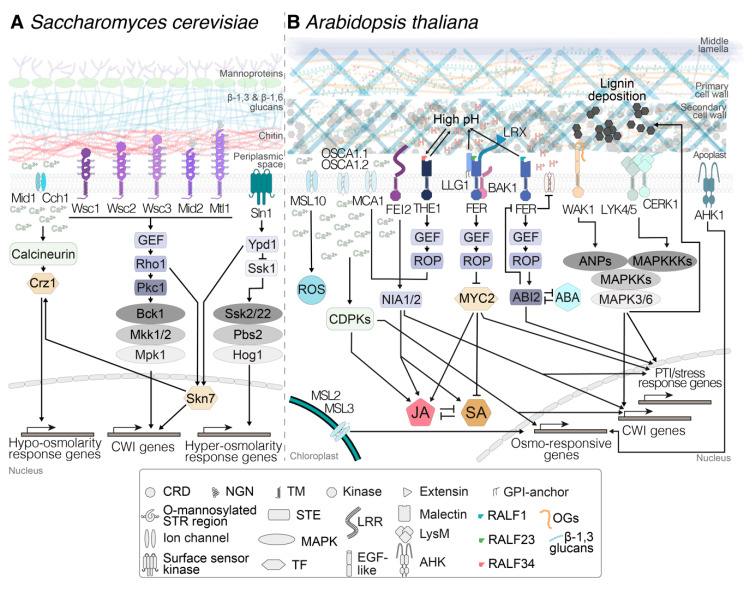
Figure 1.
Comparison between cell wall integrity (CWI) maintenance mechanisms in (A) Saccharomyces cerevisiae and (B) Arabidopsis thaliana. In both organisms, mechanosensitive ion channels and receptors trigger signal transduction processes involving Ca2+ influx into the cytoplasm and the activation of cascades including calcium-dependent protein kinases (CDPKs), guanosine nucleotide exchange factors (GEFs) and mitogen-activated protein kinases (MAPKs) that eventually activate transcription factors. In A. thaliana, the processes are regulated in a more intricate manner, exemplified by the complex interconnected networks regulated by jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA). The processes enable the regulation of gene expression in a tightly controlled and highly adaptive manner, allowing specific changes in cell wall and cellular metabolism to maintain cell wall integrity. Arrows are connecting elements belonging to the same pathway. ROS: reactive oxygen species; PTI: PAMP-triggered immunity; CRD: cysteine-rich domain; NGN: N-glycosilated asparagine; TM: transmembrane domain; GPI: glycosylphosphatidylinositol; STR: serine-/threonine-rich; STE: signal transduction element; LRR: leucine-rich repeat domain; LysM: lysin-motif-containing ectodomain; RALF: rapid alkalinization factor peptide; OGs: oligogalacturonides; TF: transcription factor; EGF-like: epidermal-growth-factor-like ectodomain.