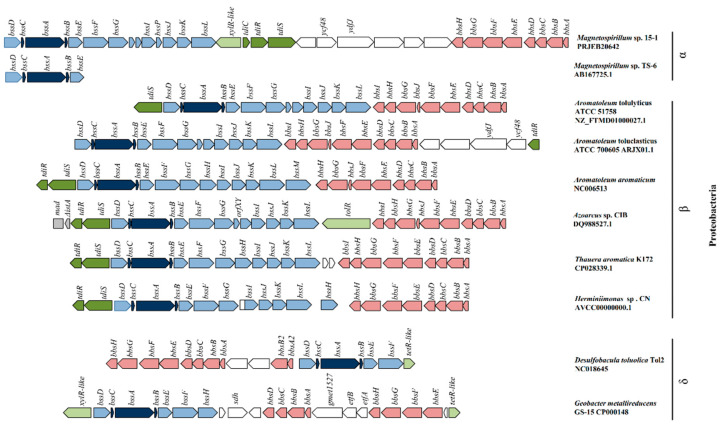
Figure 1.
Organization and comparison of the bss–bbs gene region in selected bacteria capable of anaerobic toluene degradation. Genes are represented by arrows: ‘dark blue’ for bssABC; ‘light blue’ for accessory bss genes; ‘pink’ for genes needed for transformation of benzylsuccinate to benzoyl-CoA; ‘olive’ for tdi genes; ‘light green’ for putative regulator-coding genes; ‘white’ for remaining genes without evidence for involvement in anaerobic toluene degradation. The gene names are as follows: bssABC and bssD subunits code for a benzylsuccinate synthase and its corresponding activase, respectively; bssE, putative chaperone; bssG-J and bssL, unknown function; bssK, mRNA binding protein; xylR, σ54-dependent regulator, tdiC, regulator of TdiR activity; tdiR, toluene degradation regulator; tdiS, toluene degradation inducing sensor; bbsEF, (3-methyl) benzylsuccinate CoA transferase; bbsG, (3-methyl) benzylsuccinyl-CoA dehydrogenase; bbsH, (3-methyl) phenylitaconyl-CoA hydratase; bbsCD, 2-[hydroxyphenyl(methyl)]-succinyl-CoA dehydrogenase; bbsAB, BbsAB, (3-methyl) benzoylsuccinyl-CoA thiolase.