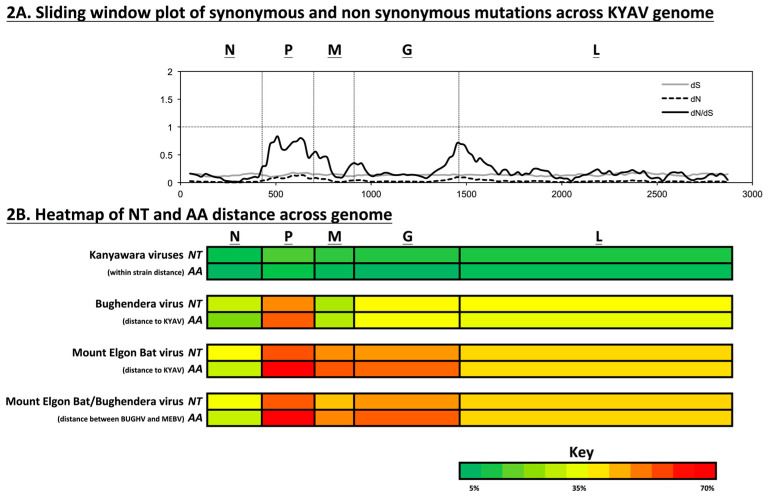
Figure 2.
Selection and diversity within KYAV-like viruses and viruses of neighbor taxa. (A) Sliding window plot (100-aa window, 20-aa step) of dN/dS across the genome of 20 KYAV-like viruses from Bundibugyo District, Uganda. Genome-wide dN/dS <1, indicating stabilizing selection predominates, but this constraint is relaxed for portions of P, M, and L. (B) Average pairwise distances of nucleotide (NT) and amino acid (AA) sequences for each virus gene within the KYAV isolates, between KYAV isolates and BUGV, between KYAV isolates and Mount Elgon bat virus (MEBV), and between BUGV and MEBV are represented by a heat map. For KYAV-like viruses in the population, average pairwise distances varied across genes, and were high for a single population (Avg NT dist: N = 5.12%, P = 9.74%, M = 7.76%, G = 7.34%, L = 7.00%; Avg AA dist: N = 1.30%, P = 6.03%, M = 2.86%, G = 0.77%, L = 4.00%).