Abstract
Cytokine-induced killer (CIK) cells are a group of heterogeneous immune cells which can be isolated from human peripheral blood mononuclear cells and have demonstrated therapeutic benefit both in hematologic malignancies and solid tumors, including colorectal cancer. However, poor tumor-targeted migration has limited the clinical efficacy of CIK cell treatment. The chemokine-chemokine receptor (CK-CKR) axis serves a role in the tumor-directed trafficking capacity of immune cells. Investigating the relationship between CKR profiles on the surface of CIK cells and chemokine expression levels in the tumor microenvironment may improve CIK cell therapy. In the present study, the spectrum of chemokine expression levels in tumor tissues from patients with colorectal cancer (CRC) and CKR expression profiles in CIK cells obtained from the same individuals with CRC were investigated. The results showed that chemokine expression levels in tumor tissues exhibited variability and cell line heterogeneity. However, the expression levels of a number of chemokines were similar in different CRC donors and cell lines. Expression levels of CXCLL10, CXCL11 and CCL3 were significantly higher in most tumor tissues compared with adjacent normal tissues and highly expressed in most CRC cell lines. In accordance with chemokine expression levels, CKR profiles on the surface of CIK cells also showed donor-to-donor variability. However, concordant expression profiles of CKRs were identified in different patients with CRC. CXCR3 and CXCR4 were highly expressed on the surface of CIK cells through the culture process. Importantly, the expression levels of all CKRs, especially CCR4, CXCR4 and CXCR3, were notably decreased during the course of CIK cell expansion. The changing trend of CKR profiles were not correlated with the chemokine expression profiles in CRC tissues (CCL3, CXCL12 and CXCL10/CXCL11 were highly expressed in CRC tissue). Re-stimulating CIK cells using chemokines (CCL21 and CXCL11) at the proper time point increased corresponding CKR expression levels on the surface of CIK cells and enhance tumor-targeted trafficking in vitro. These results demonstrated that modification of the CK-CKR axis using exogenous recombinant chemokines at the proper time point enhanced CIK cell trafficking ability and improved CIK antitumor effects.
Keywords: colorectal cancer, immunotherapy, chemokine, chemokine receptors, cytokine induce killer cells
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and ranked the second leading cause of cancer-associated mortalities worldwide in 2018 (1). Current treatment methods targeting CRC primarily include surgery assisted by radiotherapy and chemotherapy; however, high metastasis and recurrence rates remain primary causes of the high mortality associated with CRC (2). Cancer immunotherapy has received much attention and has become the focus of cancer therapy. Cytokine-induced killer (CIK) cell therapy has demonstrable therapeutic benefits, preventing cancer recurrence, increasing the quality of life of patients with cancer and extending the progression-free survival period; therefore, it has been extensively studied and applied in cancer therapies (3).
CIK cells are a group of heterogeneous cells obtained from the coculture of human peripheral blood mononuclear cells (PBMCs) and various cytokines, such as CD3McAb, interleukin (IL)-2, interferon (INF)-γ and IL-1α (4). CIK cells expressing CD3 and CD56 membrane proteins function as natural killer (NK) T cells. These CIK cells possess both the strong antitumor activity of T lymphocytes and the non-major histocompatibility complex restricted tumor-killing activity of NK cells (5–8). CIK cells can specifically target the tumor and this may be associated with the corresponding association between the chemokine expression profile of cancer cells and the chemokine receptor (CKR) expression profile in CIK cells (9).
Chemokines have similar structures and functions and their molecular weights primarily range from 8–14 kDa. Chemokines stimulate chemotactic functions in several cell types, including neutrophils, lymphocytes and monocytes (10,11). During the antitumor immune response of the body, chemokines can mediate the targeted migration of immune cells in the blood and lymph nodes to tumor locations to function in the tumor immune response. This targeted migration has become a new focus of research (12).
The present study investigated the concordance between the chemokine expression profiles of tumor tissues from patients with CRC and the CKR expression profiles of the surface of CIK cells obtained from the peripheral blood from patients with CRC. The present study aimed to increase the tumor-targeted migration ability of CIK cells through regulation of the chemokine-CKR axis in CRC.
Materials and methods
Clinical tumor and normal tissue samples
Tissue samples were collected from a total of 36 patients with histologically confirmed CRC at The Affiliated Hospital of Kunming University of Science and Technology, China. Tumor stage was classified according to the 7th edition of the Union for International Cancer Control (UICC)/American Joint Committee on Cancer TNM staging system for CRC (13). Among the 36 patients, 24 patients diagnosed with stage I–II and 12 patients diagnosed with stage III–IV. Tumor tissue and the corresponding normal tissue were collected at the same time from 7 patients who underwent surgical resection. The remaining 29 patients underwent digestive endoscopic resection and only tumor tissues were collected (Table I). Fresh tissues were washed with RPMI-1640 medium within 30 min of removal (HyClone; GE Healthcare Life Sciences) to remove traces of blood and the samples were cut into pieces for RNA extraction immediately, or stored in liquid nitrogen at −196°C. Blood samples from 20 patients with CRC (male:female, 12:8; median age ± standard deviation, 57±19 years) and 10 from healthy donors (male:female,5:5; median age ± standard deviation, 55±10 years) were also collected to cultivate CIK cells (Table I). The present study was approved by The Ethics Committee of the First People's Hospital of Yunnan Province and all patients provided written informed consent.
Table I.
Clinicopathological characteristics of patients with CRC and healthy control individuals.
| Sample used for chemokine detection | Sample used for CIK cell culture and chemokine receptor detection | ||
|---|---|---|---|
| Characteristic | Patients with CRC (n=36) | Patients with CRC (n=20) | Healthy control (n=10) |
| Age, n (%) | |||
| ≤60 years | 20 (55.6) | 11 (55.0) | 6 (60.0) |
| >60 years | 16 (44.4) | 9 (45.0) | 4 (40.0) |
| Sex, n (%) | |||
| Male | 18 (50.0) | 12 (60.0) | 5 (50.0) |
| Female | 18 (50.0) | 8 (40.0) | 5 (50.0) |
| Tumor site, n (%) | |||
| Colon | 14 (38.9) | 6 (30.0) | – |
| Rectum | 22 (61.1) | 14 (70.0) | – |
| UICC stage, n (%) | |||
| I–II | 24 (.66.7) | 12 (60.0) | – |
| III–IV | 12 (33.3) | 8 (40.0) | – |
| Histologic grade, n (%) | |||
| Well | 1 (2.8) | 0 (0.0) | – |
| Moderate | 24 (66.7) | 14 (70.0) | – |
| Poor | 11 (30.5) | 6 (30.0) | – |
| Tumor size, cm, n (%) | |||
| ≤5 cm | 30 (83.3) | 16 (80.0) | – |
| >5 cm | 6 (116.7) | 4 (20.0) | – |
| Lymph node metastasis, n (%) | |||
| None | 32 (88.9) | 18 (90.0) | – |
| Present | 4 (11.1) | 2 (10.0) | – |
CRC cell lines
The CRC cell lines DLD1, HCT116, SW480, SW620, HT29, LOVO and LS174T were purchased from the Typical Culture Preservation Committee Kunming Cell Bank, Kunming Institute of Zoology, Chinese Academy of Sciences. All of the cell lines were cultured in RPMI-1640 (Hyclone; GE Healthcare) containing 10% fetal bovine serum (FBS; Biological industries) and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin, Biological Industries) in a humidified incubator with 5% CO2 at 37°C.
RNA extraction and detection of chemokine expression profiles in tissues and cells using RT-qPCR
Total RNA was extracted from tissue samples and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration and purity of total RNA samples were verified using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into cDNA using a GoScriptTM Reverse Transcription system (Promega Corporation) according to the supplied instructions. In brief, 5 µg RNA was mixed with the primers and nuclease-free water completely and heated in a 70°C heat block for 5 min. The RNA was then placed in ice water for ≥5 min. Then the reverse transcription reaction mixture was prepared. The reverse transcription mix was combined with 5 µl of RNA and primer mix and the following temperature protocol were used: Annealing at 25°C for 5 min, extension at 42°C for 1h and then inactivation of reverse transcriptase at 70°C for 15 min. The cDNA was then used to measure the expression levels of CCL3, CCL5, CCL17, CCL19, CCL21, CCL22, CXCL10, CXCL11 and CXCL12 in CRC tumor and adjacent samples, as well as in CRC cells using SYBR Fast qPCR Master mix (Kapa Biosystems; Roche Diagnostics) and a Roche LightCycler 480 (Roche Diagnostics). The sequences of the primers used for RT-qPCR are shown in Table II (Takara Biotechnology Co., Ltd.). GAPDH was used for data normalization or reference of chemokines expression level in the CRC cell lines. The qPCR mixture was comprised of 10 µl 2× KAPA SYBR FAST qPCR Master mix, 0.4 µM each primer, 0.5 µl cDNA and PCR-grade water in a final reaction volume of 20 µl. The thermocycling conditions were as follows: Pre-incubation at 95°C for 3 min, amplification at 95°C for 10 sec, 56°C for 20 sec and extension/data acquisition at 72°C for 30 sec, for a total of 40 cycles. The melt curve was as follows: 95°C for 5 sec, 65°C for 1 min, with a continuous increase in temperature from 65–97°C at the rate of 0.02°C/sec with 10 signal acquisitions per degree; and cooling at 40°C for 10 sec. The 2−ΔΔCq method was used for data analysis (14).
Table II.
Primers used for reverse transcription-quantitative PCR.
| Gene | Sequence, 5′→3′ |
|---|---|
| GAPDH | |
| Forward | TGACTTCAACAGCGACACCCA |
| Reverse | CACCCTGTTGCTGTAGCCAAA |
| CCL3 | |
| Forward | TGTTGCCAAACAGCCACAC |
| Reverse | CAGAGCAAACAATCACAAACACAC |
| CCL5 | |
| Forward | TCCCACAGGTACCATGAAGGTC |
| Reverse | GCAATGTAGGCAAAGCAGCAG |
| CCL17 | |
| Forward | CAGAGGGACCTGCACACAGA |
| Reverse | TTCAGCTTTCTAAGGGGAATGG |
| CCL19 | |
| Forward | ACCAGGCTTCCAGCTCCTCT |
| Reverse | ACCAGGCGGCTTTATTGGT |
| CCL21 | |
| Forward | GCCACACTCTTTCTCCTGCTTT |
| Reverse | ACTCTCCCTCCTCGGTCTCTCT |
| CCL22 | |
| Forward | GGTATTTGAACCTGTGGAATTGGAG |
| Reverse | CAGGCCCTGGATGACACTGA |
| CXCL10 | |
| Forward | TGCAAGCCAATTTTGTCCAC |
| Reverse | GACCTTTCCTTGCTAACTGCTTTC |
| CXCL11 | |
| Forward | GCTGTGATATTGTGTGCTACAGTTG |
| Reverse | TTGGGTACATTATGGAGGCTTTC |
| CXCL12 | |
| Forward | CCCTGCTTACCCGCAAAA |
| Reverse | CTTCAGAGGCAATCACAAAACC |
Extraction of CIK cells
Blood samples from healthy donors or patients with CRC were processed using Ficoll-Hypaque density gradient centrifugation (Beijing Solarbio Science and Technology Co., Ltd.) to obtain PBMCs. After washing in RPMI-1640 medium, 2×106 cells were resuspended in RPMI-1640 medium containing 10% FBS, 2 mM glutamine, 100 IU/ml penicillin and 100 IU/ml streptomycin in a cell culture flask. After incubation for 24 h with 1,500 IU/ml INF-γ (Shanghai ChemoWanbang Biopharma Co., Ltd.; cat. no. S10980084), 5,000 IU/ml IL-2 (Beijing T&L Biotechnology Co., Ltd; cat. no. TL-104) and 100 ng/ml anti-CD3 antibodies (1:10,000; Wuhan Institute of Biological Products Co., Ltd.; cat. no. S19990012) were added and maintained at 37°C in humidified atmosphere of 5% CO2 for 2 days. Then fresh culture medium containing 5,000 IU/ml recombinant human (rh)IL-2 was added every 2 to 3 days. In some assays, the recombinant chemokine ligand CCL21 (100 ng/ml) or CXCL11 (10 ng/ml) (PeproTech, Inc.) was added during the culture process. The cells were maintained at 37°C in a humidified atmosphere of 5% CO2. After 13 days, cells were harvested for flow cytometry or Transwell analysis.
Detection of the targeted migration ability of CIK cells using a Transwell assay
To assess the effects of CRC cells with different expression levels of chemokines on the targeted migration ability of CIK cells, DLD1 (low expression of chemokine CCL21 and CXCL11), SW480 (high expression of chemokine CCL21) and HT29 (high expression chemokine of CXCL11) were used. In brief, 200 µl CIK cell suspension containing 5×105 cells was inoculated in the top chamber of a 24-well Transwell plate (5 µm; Corning Inc.) and 600 µl CRC cell suspension (DLD1, SW480 or HT29) containing 5×105 cells was added to the bottom chamber and placed in an incubator at 37°C with 5% CO2 for 24 h. Then the number of CIK cells that had migrated to the bottom chamber was imaged and counted using a flow cytometer. To assess the effects of different concentrations of recombinant human chemokine proteins on the targeted migration ability of CIK cells, 200 µl CIK cell suspension containing 5×105 cells was inoculated in the top chamber of a Transwell plate and 600 µl of RPMI-1640 contained 10% FBS and either rhCCL21 (final concentration 100 or 10 ng/ml) or rhCXCL11 (final concentration 10 or 0.1 ng/ml) was added to the bottom chamber. To assess the effects of the chemokine pretreatment of CIK cells on the expression levels of CKRs and cell-targeted migration ability, after culturing the CIK cells for 12 days, either rhCCL21, (final concentration of 100 ng/ml) or rhCXCL11 (final concentration 10 ng/ml) was added to the CIK cell culture system. After culturing the cells for another 2 days, 200 µl CIK cell suspension containing 5×105 cells was collected and inoculated in the top chamber of a Transwell plate and 600 µl of RPMI-1640 containing 10% FBS and either rhCCL21 (final concentration 100 ng/ml) or rhCXCL11 (final concentration 10 ng/ml) was added to the bottom chamber. The control was composed of CIK cells without pretreatment with the recombinant chemokine. Cells were placed in an incubator at 37°C with 5% CO2 for 24 h. After the top chamber of the Transwell was discarded, the number of CIK cells that had migrated to the bottom chamber was imaged and counted using a flow cytometer.
Detection of the CKR expression profiles in CIK cells using flow cytometry
For CIK cells harvested on days 7, 14, 21 and 28 were washed twice with washing buffer (PBS buffer containing 0.5% BSA), blocked with blocking buffer (PBS containing 2% BSA) for 10 min at 4°C and washed twice again. Then the washed CIK cells were stained with 5 µl of the following monoclonal antibodies in 100 µl blocking buffer (PBS containing 1% BSA) for 30 min at 4°C, washed twice, resuspended in 100 µl PBS buffer and analyzed via flow cytometry. The following antibodies were used and diluted 1:20 in blocking buffer (PBS containing 1% BSA): CD3-FITC (cat. no. 11-0036-42), CD56-APC (cat. no. 17-0567-42), CCR4-PE (cat. no. 12-1949-41), CCR5-PE (cat. no. 12-1956-41), CCR7-PE (cat. no. 12-1979-42), CXCR3-PE (cat. no. 12-1839-42) and CXCR4-PE (cat. no. 12-9999-41) antibodies, as well as the corresponding isotype controls IgG2a-PE (cat. no. 12-4321-80), IgG1a-PE (cat. no. 12-4714-82), IgG1a-FITC (cat. no. 11-4714-81) and IgG1a-APC (cat. no. 17-4714-82). All antibodies were purchased from eBioscience; Thermo Fisher Scientific, Inc. Data were obtained using a MoFlo flow cytometer (BeckmanCoulter, Inc.) and analyzed using Summit version 5.2 (Beckman Coulter, Inc.) and FlowJo version 10 software (Becton, Dickinson and Company).
Statistical analysis
GraphPad software version 5.0 (GraphPad Software) was used for statistical analysis. Analyses were performed using an unpaired Student's t-test with Welch's correction and one-way or two-way ANOVA with Bonferroni's correction or Tukey-Kramer post-hoc tests where appropriate. The statistical test used for each figure is described in the corresponding figure legend. P<0.05 was considered to indicate a statistically significant difference.
Results
Different levels of chemokine expression were detected in CRC tissues
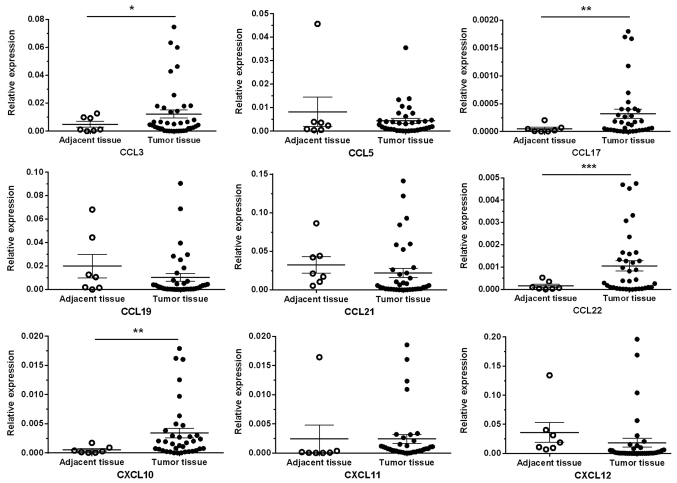
The RT-qPCR results revealed significantly higher expression levels of the chemokine ligands CCL3, CCL17, CCL22 and CXCL10 in cancer tissues (n=36) compared with adjacent normal tissues (n=7) (P<0.05, P<0.01, P<0.001 and P<0.01, respectively), whereas other chemokine ligands (CCL5, CCL19, CCL21, CXCL11 and CXCL12) exhibited no significant differences between CRC tumor tissues and adjacent normal tissues (Fig. 1).
Figure 1.
Chemokine expression profiles of CRC tumor and adjacent tissues. The expression levels of chemokine ligands CCL3, CCL17, CCL22 and CXCL10 in cancer tissues (n=36) was significantly higher compared with distant normal tissues (n=7). Meanwhile, the expression levels of other chemokine ligands (CCL5, CCL19, CCL21, CXCL11 and CXCL12) had no significant differences between CRC tumor sites and adjacent normal tissue. *P<0.05; **P<0.01; ***P<0.001. The comparisons were performed by unpaired Student's t-test with Welch's correction. CRC, colorectal cancer.
Chemokine expression profiles in different patients with CRC and CRC cell lines showed both common characteristics and donor-to-donor variation
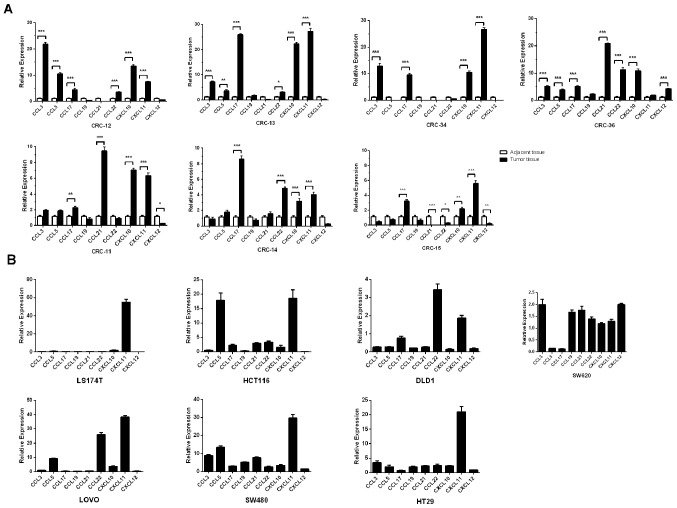
The chemokine expression levels were evaluated in both the colon tumor tissues and corresponding adjacent normal tissues of 7 patients with CRC. In addition, 7 CRC cell lines (174T, HCT116, DLD1, SW620, LOVO, SW480 and HT29) were also evaluated. The data demonstrated that the expression levels of certain chemokines were concordant in the cancer tissues of different patients with CRC and different CRC cell lines. For example, CXCL11 expression levels were significantly higher in 6/7 cases of cancer tissues from patients with CRC compared with normal tissues (all P<0.001; Fig. 2A) and highly expressed in 5/7 CRC cell lines chemokines were upregulated more than 5-fold compared with GAPDH (Fig. 2B). Similarly, the expression levels of CCL3 were significantly higher in 4/7 cancer tissues compared with adjacent normal tissues (P<0.001; Fig. 2A) and 3/7 CRC cell lines relative to GAPDH expression (>2-fold of the CCL3/GAPDH expression ratio was the cut off value used. The expression ratio of CCL3/GAPDH in SW620, SW480 and HT29 was 2.00±0.15, 8.85±0.35 and 3.45±0.39, respectively; Fig 2B). In addition, CXCL10 expression levels were higher in 7/7 cases of CRC compared with adjacent normal tissue (CRC tissue vs. adjacent normal tissue; 6/7 samples P<0.001 and 1/7 sample P<0.01) and 3/7 CRC cell lines (LOVO, SW480 and HT29) compared with GAPDH exhibited a 2-fold change in the CCL3/GAPDH expression ratio. The ratio of CXCL10/GAPDH in LOVO, SW480 and HT29 was 3.69±0.23, 3.42±0.37 and 2.41±0.09, respectively. The expression levels of CCL19 did not differ between all the cancer tissues and normal tissues. The same result was also observed in 6/7 CRC cell lines (with the exception of SW480). However, the chemokine expression profiles in cancer tissues from different patients with CRC and in different CRC cells showed heterogeneity (Fig. 2A and B).
Figure 2.
Chemokine expression profiles of patients with CRC and CRC cell lines. The expression levels of 9 chemokine ligands were detected in tumor and the paired adjacent normal samples from (A) 7 patients with CRC(CRC-11, CRC-12, CRC-13, CRC-14, CRC-15, CRC-34 and CRC-36) and (B) 7CRC cell lines (DLD1, HCT116, SW620, SW480, HT29, LOVO and LS174T) using reverse transcription-quantitative PCR. The expression levels of 3 chemokine ligands (CXCL11, CCL3 and CCL19) were consistently upregulated in the majority of the CRC patients compared with the adjacent normal samples and also in CRC cell lines relative to the house-keeping gene GAPDH. Expression levels of CCL19 were not significantly different in tumor vs. paired adjacent normal samples and CRC cell lines relative to the house-keeping gene GAPDH. However, the expression profiles of chemokines between patients with CRC and CRC cell lines were heterogeneous. The comparisons were performed using two-way ANOVA analysis and Bonferroni's correction. *P<0.05; **P<0.01; ***P<0.001. CRC, colorectal cancer.
CXCR3 and CXCR4 were more highly expressed on the surface of CIK cells derived from patients with CRC compared with those derived from healthy controls
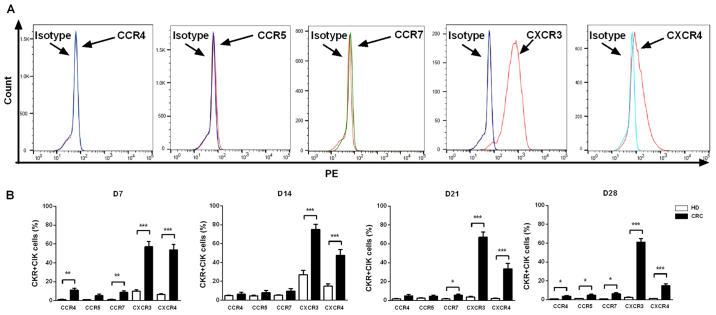
On days7, 14, 21 and 28 of culture of peripheral blood CIK cells from patients with CRC (n=20) and healthy individuals (n=10), CKR expression levels in CIK cells were detected using flow cytometry. The results indicated that the CIK cells derived from patients with CRC or healthy individuals showed similar CKR expression profiles. CXCR3 and CXCR4 had higher expression levels on CIK cells compared with isotype, while the expression profiles of CCR4, CCR5 and CCR7 did not show significant changes compared with isotype (Fig. 3A). At the 4 time points examined, the expression levels of both CXCR3 and CXCR4 were significantly higher in CIK cells from patients with CRC compared with those from healthy individuals (P<0.001). However, the expression levels of CCR4, CCR5 and CCR7 in CIK cells from patients with CRC and healthy individuals differed over time; CCR4 expression levels on day 7 (P<0.01) and day 28 (P<0.05), CCR5 on day 28 (P<0.05) and CCR7 on day 7 (P<0.01), 21 (P<0.05) and 28 (P<0.05) were significantly higher in CIK cells from patients with CRC compared with those from healthy individuals (Fig. 3B).
Figure 3.
Expression levels of chemokine receptors on CIK cells generated from patients with CRC and healthy donors. (A) Analysis of the expression levels of CCR4, CCR5, CCR7, CXCR3 and CXCR4 on CIK cells via flow cytometry revealed that CXCR3 and CXCR4 had higher expression levels on CIK cells compared with isotype, expression profiles of CCR4, CCR5 and CCR7 did not show significant changes compared with isotype. (B) Dynamic changes of chemokine receptors CCR4, CCR5, CCR7, CXCR3 and CXCR4 of CIK cells on D7, 14, 21 and 28 were detected in patients with CRC and HD CIK cells via flow cytometry. The result revealed that expression levels of CXCR3 and CXCR4 were significantly higher on CIK cells cultured from CRC compared with HD at all the time points analyzed, while the expression levels of CCR4, CCR5 and CCR7 were significantly higher on CIK cells cultured from CRC compared with HD at different time points analyzed (CCR4 on D7 and D28; CCR5 on D28; CCR7 on D7, D21 and D28). All comparisons were performed using unpaired Student's t-test with Welch's correction. *P<0.05; **P<0.01; ***P<0.001. CRC, colorectal cancer; HD, healthy donors; CIK, cytokine-induced killer cell.
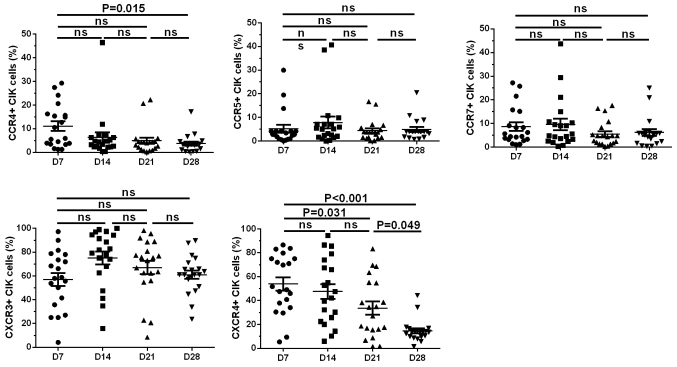
Expression levels of CKRs on the surface of CIK cells decreased gradually during the expansion process
Analyses of the changing trends of the CKR expression profiles of CIK cells from patients with CRC (n=20) over time showed that CCR4 and CXCR4 expression levels peaked on day 7 (11.06±2.020 and 53.89±5.539%, respectively); then, CCR4 and CXCR4 expression levels gradually decreased and reached their minimum values on day 28 (3.697±0.839 and 14.630±2.112%, respectively). The changes in expression levels of day 28 and day 7 in CCR4 and CXCR4, respectively were significant (P=0.015 for CCR4 and P<0.001 for CXCR4). Expression levels of CCR5, CCR7 and CXCR3 were detected on day 7 (5.165±1.673, 8.663±1.810 and 57.040±5.486%, respectively) and then these expression levels reached their peak on day 14 (7.855±2.521, 9.620±2.410 and 75.140±5.319%, respectively) and then gradually decreased. On day 28, the expression levels of CCR5, CCR7 and CXCR3 returned to levels similar to those on day 7 (4.913±1.006, 6.209±1.415 and 61.060±3.577%, respectively); however, the changes in expression levels of these 3 CKRs over time were not significant (Fig. 4).
Figure 4.
CKR expression level trends over time in culture. Levels of each CKR are shown at D7, 14, 21 and 28 for each CRC expansion. The expression levels of CCR4 and CXCR4 on the CIK cells peaked on D7 and gradually reduced over time in culture. A trend towards significantly reduced CKR expression levels was observed over the time points for CCR4 (D28 vs. D7: P=0.015) and CXCR4 (D28 vs. D7: P<0.001, D28 vs. D21: P=0.049 and D21 vs. D7: P=0.031). However, the changes in expression levels of these CCR5, CCR7 and CXCR3 over time were not significant. Comparisons were performed using one-way ANOVA with Bartlett's correction. CKR, chemokine receptor; CRC, colorectal cancer; CIK, cytokine-induced killer cell; D, day.
CKR expression levels could be boosted by adding exogenous recombinant chemokines
CCL21 and CXCL11 were separately added to the CIK cell culture system on days 5, 12, 19 and 26. After culturing for another 2 days (to days 7, 14, 21 and28, respectively), the CIK cells were collected and the expression levels of CCR7 (the corresponding ligand of CCL21) and CXCR3 (the corresponding ligand of CXCL11) (15) were detected using flow cytometry. CCR7 expression levels on day 14 were significantly higher in CIK cells pretreated with CCL21 compared with CIK cells without pretreatment (8.350 vs. 4.235%; P<0.05), whereas CCR7 expression levels were not significantly increased at the 3other time points. CXCR3 expression levels were significantly higher on day 7 in CIK cells pretreated with CXCL11 compared with CIK cells without pretreatment (35.500 vs. 9.570%; P<0.05), whereas CXCR3 expression levels were not significantly increased at the 3 other time points (Fig. 5).
Figure 5.
Expression of chemokine receptors on CIK cells could be elevated by corresponding recombinant chemokines. Chemokine ligands CCL21 and CXCL11 were added to the CIK culture process respectively to stimulate the expression level of corresponding chemokine receptors CCR7 and CXCR3 on CIK cells. As data showed that compared with the untreated group the percentage of CCR7(+) CIK cells was significantly enhanced after stimulation on D14. Similarly, chemokine ligand CXCL11 elevated the percentage of CXCR3(+) CIK cells after stimulation on D7. Comparisons were performed using two-way ANOVA analysis and Bonferroni's correction *P<0.05. CIK, cytokine-induced killer cells; D, day.
Boosting CKR expression levels enhances the tumor-targeted trafficking ability of CIK cells
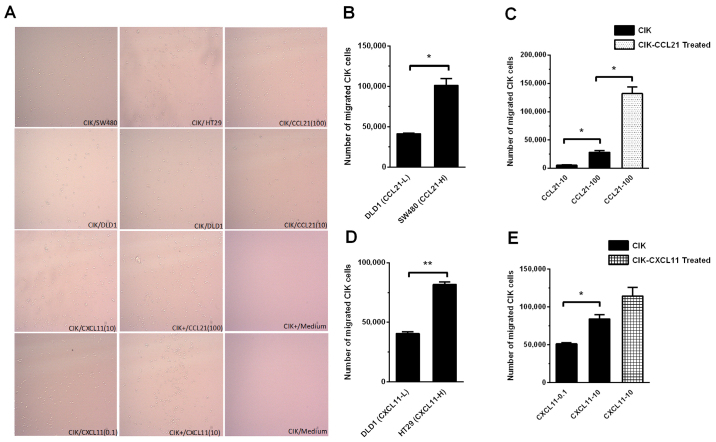
To explore the trafficking ability of CIK cells, a Transwell assay was performed and the CIK cells that migrated into the lower chamber were observed under a microscope (Fig. 6A) or counted using flow cytometry (Fig. 6B-E). When the bottom chamber of the Transwell was inoculated with DLD1 cells with low CCL21 expression levels (CCL2-L) and SW480 cells with high CCL21 expression levels (CCL2-H), DLD1 cells had a significantly reduced effect on the chemotactic ability of CIK cells compared with that of SW480 cells (41,430±575 vs. 101,300±8,250; P<0.05; Fig. 6B). Similar results were obtained in experiments using different concentrations of recombinant CCL21 to influence the chemotactic ability of CIK cells. When the CCL21 concentrations in the bottom chamber of the Transwell were 10 ng/ml and 100 ng/ml, 10 ng/ml CCL21 had a significantly lower effect on the chemotactic ability of CIK cells compared with 100 ng/ml CCL21 (5,400±900 vs. 28,450±3,050; P<0.05; Fig. 6C). When the CCL21 concentration in the bottom chamber of the Transwell was 100 ng/ml, a significantly greater number of CIK cells cultured to day 14 migrated from the top to the bottom chamber pretreated with exogenous CCL21 vs. without CCL21 pretreatment (132,000±12,000 vs. 28,450±3,050; P<0.05; Fig. 6C).
Figure 6.
Chemokine ligand-treated CIK cells showed greater migration in Transwell assay. (A) Treated or untreated CIK cells were added into the upper well and recombinant chemokine or tumor cells were added into the lower well of Transwell chamber. Numbers of migrated immune cells were imaged and counted using flow cytometry. (B) CRC cell lines with different chemokine ligand expression levels showed different abilities of CIK cell migration. CIK cells exhibited higher ability to migrate to SW480 (CCL21-H) compared with DLD1 (CCL21-L). (C) CIK cells exhibited higher ability to migrate with a higher concentration of recombinant CCL21 (100 ng/ml) compared with lower concentration of CCL21 (10 ng/ml). When the concentration of recombinant CCL21 was the same (100 ng/ml), CIK cell pre-treated with CCL21 exhibited higher ability to migrate to the lower chamber compared with untreated CIK cells. (D) CIK cells exhibited higher ability to migrate to HT29 (CXCL11-H) than DLD1 CXCL11-L). (E) CIK cells exhibited higher ability to migrate to higher concentration of recombinant CXCL21 (10 ng/ml) compared with lower concentration of CXCL21 (0.1 ng/ml). When concentration of recombinant CXCL21 was the same (10 ng/ml), CIK cell pre-treated by CXCL21 exhibited no significant ability to migrate to the lower chamber when compared with untreated CIK cells. Unpaired Student's-tests were performed to analyze the data of (B) and (D). One-way ANOVA and Tukey-Kramer post-hoc test was performed to analyze the data of (C) and (E). *P<0.05; **P<0.01. CIK, cytokine-induced killer cells; L, low; H, high.
When DLD1 cells with low CXCL11 expression levels (CXCL11-L) and HT29 cells with high CXCL11 expression levels (CXCL11-H) were separately inoculated in the bottom chamber in the Transwell assay, DLD1 cells had a significantly reduced effect on the chemotactic ability of CIK cells compared with HT29 cells (40,500±1,500 vs.79,250±2,750; P<0.01; Fig. 6D). Similar results were also obtained using different concentrations of recombinant CXCL11 to influence the chemotactic ability of CIK cells. The addition of 0.1 ng/ml CXCL11 to the bottom chamber had a significantly reduced effect on the chemotactic ability of CIK cells compared with of 10 ng/ml CXCL11 (51,250±1,750 vs. 84,000±600; P<0.05; Fig. 6E). At a CXCL11 concentration of 10 ng/ml in the bottom chamber, there was no significant increase in the number of CIK cells that migrated to the bottom from the top chamber pretreated with exogenous CXCL11 and cultured to day 14 vs. without CXCL11 pretreatment (114,500±11,500 vs. 84,000±6,000, respectively; P>0.05) (Fig. 6E).
Discussion
Due to their strong cell-killing capacity and broad antitumor spectrum, CIK cells can effectively kill chemotherapy-resistant tumor cells. CIK cells exhibit clear efficacy regarding the clearance of minimal residual lesions of tumors and have few side effects; therefore, CIK cells have become an extensively used treatment for adoptive cellular immunotherapy (16).
CRC is a common malignant tumor with global high morbidity and mortality rates. Most patients with CRC are already in the late stage at the time of diagnosis and because late-stage tumor cells have already spread or metastasized in patients with late-stage CRC, conventional surgery have little therapeutic benefit. In addition, patients may not be able to tolerate traditional radio-or chemotherapy; therefore, CIK cell treatment may improve the quality of life of these middle- to late-stage patients and extend patient survival time (17,18). In the clinic, however, patients with CRC who have received CIK cell treatment usually present with different clinic efficacies, whereas a number of patients do not experience any therapeutic benefit (3).
How to effectively promote the infiltration of immune-reactive cells into tumors is an important focus of cancer research in order to increase the efficacy of antitumor immune-therapies. Successful antitumor immunotherapy enhances the tumor-targeted trafficking ability of immune cells but requires effective methods for in vitro activation of these cells (19).
Chemokines are important regulatory factors that direct immune-reactive cells to tumors. By binding to corresponding CKRs on the target cell membrane, chemokines can induce the targeted migration of target cells (20). The interaction between chemokines released in an abnormal cancer microenvironment and CKRs on the surface of CIK cells is an important factor that affects the tumor-targeted migration ability of CIK cells (21). The concordance between these two variables directly affects the treatment effects of CIK cells (22–24). Currently, however, the CKR expression profiles of CIK cells derived from patients with autologous CRC and the underlying molecular mechanisms influencing the tumor-targeted migration ability of CIK cells via CKR expression profiles remain unclear. In the present study, the chemokine expression profiles in tumor tissues from patients with CRC and the CKR expression profiles of the surface of CIK cells derived from the same donors were detected. The corresponding association between these profiles was analyzed to understand the effects of the concordance between the chemokine expression profiles in the cancer microenvironment and the CKR expression profiles on CIK cells on the tumor-targeted migration ability of CIK cells. In addition, a simple modification of the chemokine-CKR axis for increasing the tumor-targeted migration ability of CIK cells was also investigated.
First, the chemokine expression profiles in CRC tissues and adjacent normal tissues were examined, as well as in CRC cell lines. The results reported that the chemokine expression profiles in the tumor tissues from patients with CRC and different CRC cell lines had common characteristics. CXCL10, CXCL11 and CCL3 were overexpressed in most CRC tumor samples and CRC cell lines. In contrast, CCL19 was expressed at low levels in both the tumor and adjacent normal samples. This effect might be due to the regulation of cancer cell proliferation and invasion through the interaction with CXCR3 and CXCR7 (25). However, the chemokine expression profiles of tumor tissues from patients with CRC and in different CRC cell lines had significant heterogeneity. Due to the fact that the different CRC cell lines were derived from CRC tumor tissues from different patients with CRC, the high heterogeneity of chemokine expression profiles in patients with CRC may be associated with pathological factors, such as tumor stage and metastasis. Whether this heterogeneity affects the response to CIK cell treatment requires evidence from clinical observational data obtained from large sample sizes.
Next, the present study examined the CKR expression profiles on the surface of CIK cells from patients with CRC and healthy individuals. The dynamic changes in CKR expression levels in CIK cells during the expansion process were also monitored. The results showed that CIK cells from the two sources had similar CKR expression profiles: CXCR3 and CXCR4 were notably more highly expressed on the surface of CIK cells compared with CCR4, CCR5 and CCR7. In addition, the CKR expression levels on the surface of CIK cells derived from patients with CRC were significantly higher compared with those derived from healthy individuals. The differences in expression levels of CXCR3 and CXCR4 were the most notable. These results were inconsistent with those of our previous work (26), which demonstrated that the increased expression levels of CKR on the surface of CIK cells did not differ significantly between patients with CRC and healthy individuals. It was hypothesized that certain factors, such as the culture condition, ethnicity and pathological stage, might be associated with these differences. The present results also differed from those described by Wang et al (9), who reported a reduction in the expression levels of CKR on the surface of CIK cells in patients with CRC compared with cells derived from healthy individuals. It was hypothesized that this discrepancy between the present study and the aforementioned study may be due to the disparate in vitro activation times of the CIK cells used for CKR detection, donor resources, such as UICC stage and other parameters. Therefore, future studies with larger sample sizes are needed.
It is noteworthy that all the CKR expression levels declined during the CIK cell culture process in both the present study and in the other two aforementioned previous reports (9,26). Therefore, due to these consistent results, the present study aimed to enhance CKR expression levels during the course of CIK cell culture and enhance CIK cell trafficking ability. Further analyses between the chemokine expression profiles in tumor tissues from patients with CRC and the CKR expression profiles on the surface of CIK cells derived from the same patients demonstrated that the chemokine and CKR expression profiles were associated. CXCR3 expression levels were higher on the surface of CIK cells and the expression of its corresponding ligand, CXCL10, was also higher in CRC tumor tissues compared with normal tissues. In addition, the expression levels of CCR4 were higher on the surface of CIK cells and the expression levels of its corresponding ligands, CCL3 and CCL22, were also higher in CRC tumor tissues compared with adjacent normal tissues. It was hypothesized that the corresponding association between chemokines and CKRs was important for allowing CIK cells to migrate to tumor tissue in patients with CRC. Consistent with the present study, Wang et al (9) demonstrated that expression levels CXCL10 was elevated in CRC tumor tissues compared with paracancerous tissues and that the expression levels of its corresponding ligand, CXCR3, were also increased in CIK cells derived from patients with CRC compared with PBMCs before activation. However, no corresponding association between chemokine and CKR expression profiles was observed in the present study. For example, CXCR4 expression levels were elevated on the surface of CIK cells but the expression levels of its corresponding ligand, CXCL12, were lower in CRC tumor tissues compared with paracancerous tissues. In addition, CCR7 expression levels were higher on the surface of CIK cells, but the expression levels of its corresponding ligands, CCL19 and CCL21, were not significantly different between CRC tumor tissues and normal tissues. The discrepancy between cytokine expression levels from tumor tissue and CKRs from CIK cells might impair CIK cell tumor-targeted migration and limit the clinic efficacy of CIK cells in CRC.
The expression levels of chemokines in CRC cells partially determine the targeted migration ability of CIK cells. Several studies have indicated that chemokines induce the targeted migration of lymphocytes (27–30). Therefore, strategies to alter the local concentration of chemokines in tumors, increase the expression levels of CKR on the surface of immune cells and utilize the chemokine-CKR axis to increase tumor-targeted migration and infiltration of immune cells, such as lymphocytes, NK cells and CIK cells, into tumors may improve our theoretical understanding for treatment of cancer. For example, Pevida et al (31) added the chemokine ligands CCL3 and CCL5 to NCTC2472 mouse fibrosarcoma cells to increase the expression levels of the corresponding receptor CCR1 on the cell surface. In our previous work (26), CD3/CD28 magnetic beads were added to a CIK culture system to stimulate the expression of CCR5, CCR7 and CXCR3 on the CIK cell surface.
As aforementioned, almost all the CKRs declined during the expansion process in the present study. To boost CKR expression levels in CIK cells, 2 chemokines (CCL21 and CXCL11) were selected. The results indicated that CCL21 or CXCL11 pretreatment significantly increased the expression levels of CCR7 or CXCR3 on the CIK cell surface. These results suggest that the addition of exogenous recombinant chemokines during the culture process of CIK cells might increase the expression levels of ligands corresponding to chemokines on the surface of CIK cells. However, it was also observed that the expression levels of CCR7 were significantly higher at day 14 on the surface of CIK cells with CCL21 pretreatment vs. without CCL21 pretreatment, whereas CXCR3 expression levels were significantly higher at day 7 on the surface of CIK cells with CXCL11 pretreatment vs. without pretreatment. These results suggested that when adding exogenous recombinant chemokines to increase the expression levels of ligands corresponding to chemokines on the surface of CIK cells, different time points should be selected based on disparate chemokines.
Furthermore, the present study reported that increased CKR expression levels on the surface of CIK cells could enhance the tumor-targeted migration ability of CIK cells. A significantly greater number of CIK cells had migrated to the bottom Transwell chamber after pretreatment with CCL21 vs. without treatment. However, CIK cells cultured to day 14 pretreated with CXCL11 did not exhibit increased CXCR3 expression levels because the CXCR3 expression levels in CIK cells can be boosted significantly with CXCL11 on day7 but no statistical difference was observed at other time points. Therefore, the observed chemotactic ability was not significantly increased. These results further demonstrated that the tumor-targeted migration ability of CIK cells is chemokine-CKR dependent and that it was feasible to increase the tumor-targeted migration ability of CIK cells with chemokine pretreatment during the CIK culture process at the proper time point.
There are some limitations of the present study. CIK cells are a heterogeneous population, which compose various subsets, such as CD3(+)CD56(+) CIK, CD3(+) CD56(−) T and CD3(−)CD56(+) NK cells (32). CD3(+)CD56(+) CIK cells appear to possess the most potent cytotoxicity and high impact on adoptive cellular immunotherapy (33). However, other subtypes of CIK cells may have different immunologic functions. For example, alloreactivity against human leukocyte antigen-mismatched PBMC is restricted to CD3(+)CD56(−) CIKs (34). Therefore, the association between chemokines and CKRs from various subsets of total CIK cells should be considered in future research. Then, the relative expression levels of chemokines in CRC cell lines were compared with GAPDH as a reference in the present study. However, GAPDH at the mRNA level in CRC sample was increased significantly compared with the paired non-cancerous part (35). Therefore, it will be better to use a normal control cell line to measure differential CKR expression in CRC cell lines. Next, 2 chemokines (CCL21 and CXCL11) were selected to boost CKR expression levels during CIK cell culture process in the present study. However, whether these two chemokines affect the proliferation of CIK cells requires further study. Finally, the present study only measured chemokine expression levels in CRC and adjacent normal tissue using RT-qPCR. It would be better to perform western blots to verify the results of the present study at the protein expression level.
Taken together, the results of the study demonstrated that CXCL10, CXCL11 and CCL3 expression levels were significantly higher in CRC tumor tissues compared with adjacent normal tissue. However, the levels of all CKRs of CIK cells, especially CCR4, CXCR4 and CXCR3, decreased considerably during the course of CIK cell expansion. Notably, re-stimulating CIK cells with chemokines CCL21 at day 14 and CXCL11 at day 7 significantly increased the corresponding CKR expression levels on the surface of CIK cells and enhanced tumor-targeted trafficking in vitro. To the best of our knowledge, the present study is the first to show that, although adding exogenous recombinant chemokines to increase the impaired CKRs expression on the surface of CIK cells, different time points should be selected based on disparate chemokines. Therefore, evaluating the expression levels of chemokines in the CRC tumors and stimulating with proper exogenous recombinant chemokines at proper time point could increase corresponding CKR expression levels, enhance CIK cell tumor-targeted trafficking and improve clinic efficacy.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National Natural Science Foundation of China (grant nos. 81460463 and 81502556), The Medical Academic Talents Cultivation Foundation for Health Commission of Yunnan Province (grant no. D-201642) and The Kunming Key Laboratory of Tumor Molecular and Immune Prevention Foundation (grant no. 2018-1-A-17334).
Availability of data and materials
All the datasets generated and analyzed in the present study are included in this published article.
Authors' contributions
YZ and HT designed the study and wrote the manuscript. XY, HT and QG carried out the concepts, definition of intellectual content and reviewed the manuscript. YZ, JL and JW performed the experiments. LW and JZ collected the data. XY, DL and JF analyzed and interpreted the data. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Ethics Committee of the First People's Hospital of Yunnan Province and all patients gave written informed consent. The approval number was 2018GJ111.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal Cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 3.Hontscha C, Borck Y, Zhou H, Messmer D, Schmidt-Wolf IG. Clinical trials on CIK cells: First report of the international registry on CIK cells (IRCC) J Cancer Res Clin Oncol. 2011;137:305–310. doi: 10.1007/s00432-010-0887-7. [DOI] [PubMed] [Google Scholar]
- 4.Giancola R, Olioso P, Di Riti M, Capone A, Contento A, Pompetti F, Iacone A. Evaluation of an automated closed fluid management device for processing expanded cytokine-induced killer cells to use in immunotherapy programs for cancer. Transfusion. 2008;4:629–639. doi: 10.1111/j.1537-2995.2007.01587.x. [DOI] [PubMed] [Google Scholar]
- 5.Baker J, Verneris MR, Ito M, Shizuru JA, Negrin RS. Expansion of cytolytic CD8(+) natural killer T cells with limited capacity for graft-versus-host disease induction due to interferon gamma production. Blood. 2001;97:2923–2931. doi: 10.1182/blood.V97.10.2923. [DOI] [PubMed] [Google Scholar]
- 6.Lu PH, Negrin RS. A novel population of expanded human CD3+CD56+ cells derived from T cells with potent in vivo antitumor activity in mice with severe combined immunodeficiency. J Immunol. 1994;153:1687–1696. [PubMed] [Google Scholar]
- 7.Leemhuis T, Wells S, Scheffold C, Edinger M, Negrin RS. A phase I trial of autologous cytokine-induced killer cells for the treatment of relapsed Hodgkin disease and non- Hodgkin lymphoma. Biol Blood Marrow Transplant. 2005;11:181–187. doi: 10.1016/j.bbmt.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Verneris MR, Karimi M, Baker J, Jayaswal A, Negrin RS. Role of NKG2D signaling in the cytotoxicity of activated and expanded CD8+ T cells. Blood. 2004;103:3065–3072. doi: 10.1182/blood-2003-06-2125. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Li J, Liu JY, Li F, Wang LP, Huang L, Li JY, Chen XF, Liu JB, Wu CC, et al. Modification of chemokine receptor expression to enhance levels of trafficking receptors on autologous cytokine-induced killer cells derived from patients with colorectal cancer. Biomed Pharmacother. 2014;68:551–556. doi: 10.1016/j.biopha.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Horuk R. Chemokines. Scientific World Journal. 2007;19:224–232. doi: 10.1100/tsw.2007.6. [DOI] [Google Scholar]
- 11.Kufareva I, Salanga CL, Handel TM. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol Cell Biol. 2015;4:372–383. doi: 10.1038/icb.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Vorst EP, Döring Y, Weber C. Chemokines. Arterioscler Thromb Vasc Biol. 2015;35:e52–e56. doi: 10.1161/ATVBAHA.115.306359. [DOI] [PubMed] [Google Scholar]
- 13.Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumors. 7thed. Oxford. 2010:Wiley–Blackwell. [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCq method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Lazennec G, Richmond A. Chemokines and chemokine receptors: New insights into cancer-related inflammation. Trends Mol Med. 2010;3:133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jäkel CE, Schmidt-Wolf IG. An update on new adoptive immunotherapy strategies for solid tumors with cytokine-induced killer cells. Expert Opin Biol Ther. 2014;7:905–916. doi: 10.1517/14712598.2014.900537. [DOI] [PubMed] [Google Scholar]
- 17.Peng H, Yao M, Fan H, Song L, Sun J, Zhou Z, Du Y, Lu K, Li T, Yin A, et al. Effects of autologous cytokine-induced killer cells infusion in colorectal cancer patients: A prospective study. Cancer Bionter Radiopharm. 2017;6:221–226. doi: 10.1089/cbr.2017.2246. [DOI] [PubMed] [Google Scholar]
- 18.Kim JS, Kin YG, Park EJ, Kim B, Lee HK, Hong JT, Kim Y, Han SB. Cell-based immunotherapy for colorectal cancer with cytokine-induced killer cells. Immune Netw. 2016;2:99–108. doi: 10.4110/in.2016.16.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anandappa AJ, Wu CJ, Ott PA. Directing traffic: How to effectively drive T cells into tumors. Cancer Discov. 2020;2:185–197. doi: 10.1158/2159-8290.CD-19-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippitz BE. Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol. 2013;14:e218–e228. doi: 10.1016/S1470-2045(12)70582-X. [DOI] [PubMed] [Google Scholar]
- 21.Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, Trella E, Galati-Fournier V, Oertli D, Däster SR, et al. Gut microbiata modulate T cell trafficking into human colorectal cancer. Gut. 2018;11:1984–1994. doi: 10.1136/gutjnl-2016-313498. [DOI] [PubMed] [Google Scholar]
- 22.Zipin-Roitman A, Meshel T, Sagi-Assif O, Shalmon B, Avivi C, Pfeffer RM, Witz IP, Ben-Baruch A. CXCL10 promotes invasion-related properties in human colorectal carcinoma cells. Cancer Res. 2007;67:3396–3405. doi: 10.1158/0008-5472.CAN-06-3087. [DOI] [PubMed] [Google Scholar]
- 23.Al-Haidari AA, Syk I, Thorlacius H. HMG-CoA reductase regulates CCL17-induced colon cancer cell migration via geranylgeranylation and RhoA activation. Biochem Biophys Res Commun. 2014;1:68–72. doi: 10.1016/j.bbrc.2014.02.078. [DOI] [PubMed] [Google Scholar]
- 24.Al-haidari AA, Syk I, Jirström K, Thorlacius H. CCR4 mediates CCL17 (TARC)-induced migration of human colon cancer cells via RhoA/Rho-kinase signaling. Int J Colorectal Dis. 2013;28:1479–1487. doi: 10.1007/s00384-013-1712-y. [DOI] [PubMed] [Google Scholar]
- 25.Singh AK, Arya RK, Trivedi AK, Sanyal S, Baral R, Dormond O, Briscoe DM, Datta D. Chemokine receptor trio: CXCR3, CXCR4 and CXCR7 crosstalk via CXCL11 and CXCL12. Cytokine Growth Factor Rev. 2013;24:41–49. doi: 10.1016/j.cytogfr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou Y, Li F, Hou W, Sampath P, Zhang Y, Thorne SH. Manipulating the expression ofchemokine receptorsenhances delivery and activity of cytokine-induced killer cells. Br J Cancer. 2014;110:1992–1999. doi: 10.1038/bjc.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauro C, Fu H, Marelli-Berg FM. T cell trafficking and metabolism: Novel mechanisms and targets for immunomodulation. Curr Opin Pharmacol. 2012;12:452–457. doi: 10.1016/j.coph.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abastado JP. The next challenge in cancer immunotherapy: Controlling T-cell traffic to the tumor. Cancer Res. 2012;72:2159–2161. doi: 10.1158/0008-5472.CAN-11-3538. [DOI] [PubMed] [Google Scholar]
- 29.Franciszkiewicz K, Boissonnas A, Boutet M, Combadière C, Mami-Chouaib F. Role of chemokines and chemokine receptors in shaping the effector phase of the antitumor immune response. Cancer Res. 2012;72:6325–6332. doi: 10.1158/0008-5472.CAN-12-2027. [DOI] [PubMed] [Google Scholar]
- 30.Bryant J, Ahern DJ, Brennan FM. CXCR4 and vascular cell adhesion molecule 1 are key chemokine/adhesion receptors in the migration of cytokine-activated T cells. Arthritis Rheum. 2012;64:2137–2146. doi: 10.1002/art.34394. [DOI] [PubMed] [Google Scholar]
- 31.Pevida M, Lastra A, Meana Á, Hidalgo A, Baamonde A, Menéndez L. The chemokine CCL5 induces CCR1-mediated hyperalgesia in mice inoculated with NCTC 2472 tumoral cells. Neuroscience. 2014;259:113–125. doi: 10.1016/j.neuroscience.2013.11.055. [DOI] [PubMed] [Google Scholar]
- 32.Tingting J, Changping W, Binfeng L. Cytokine-induced killer cells promote antitumor immunity. J Transl Med. 2013;83:1–9. doi: 10.1186/1479-5876-11-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linn YC, Lau SK, Liu BH, Ng LH, Yong HX, Hui KM. Characterization of recognition and functional heterogeneity exhibited by cytokine-induced killer cell subsets against acute myeloid leukaemia target cell. Immunology. 2009;3:423–435. doi: 10.1111/j.1365-2567.2008.02910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sangiolo D, Martinuzzi E, Todorovic M, Vitaggio K, Vallario A, Jordaney N, Carnevale-Schianca F, Capaldi A, Geuna M, Casorzo L, et al. Alloreactivity and anti-tumor acitivity segregate within two distinct subsets of cytokine-induced killer (CIK) cells: Implications for their infusion across major HLA barriers. Int Immunol. 2008;20:841–848. doi: 10.1093/intimm/dxn042. [DOI] [PubMed] [Google Scholar]
- 35.Guo C, Liu S, Sun MZ. Novel insight into the role of GAPDH playing in tumor. Clin Transl Oncol. 2013;15:167–172. doi: 10.1007/s12094-012-0924-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the datasets generated and analyzed in the present study are included in this published article.
Authors' contributions
YZ and HT designed the study and wrote the manuscript. XY, HT and QG carried out the concepts, definition of intellectual content and reviewed the manuscript. YZ, JL and JW performed the experiments. LW and JZ collected the data. XY, DL and JF analyzed and interpreted the data. All the authors read and approved the final manuscript.