Table 2.
Binding energy scores and molecular interactions between top 5 leads and SARS-CoV Mpro. The figures in bracket indicate the hydrogen bond length.
| ZINC ID | Common Name | Structure | Binding Energy (kcal/mol) | Molecular Interactions |
|
|---|---|---|---|---|---|
| Hydrogen bonds | Hydrophobic interactions | ||||
| ZINC000026985532 | Saquinavir | 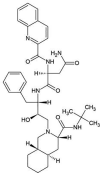 |
−9.6 | O6…..N(Glu166) [2.83 Å] N5…..OE1(Gln189) [3.06 Å] O2…..O(Leu141) [2.75 Å] O1…..N(Gly143) [2.89 Å] |
Thr25,Thr26, His41,Met49, Phe140, Asn142, Cys145, His164, Met165, Leu167, Thr190 and Gln192 (N = 12) |
| ZINC000052955754 | Ergotamine | 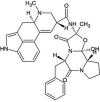 |
−9.5 | O1…..N(Gly143) O4…..N(Gly143) |
Thr25, His41, Met49, Asn142, Cys145, His164, Met165, Glu166 and Gln189 (N = 9) |
| ZINC000006716957 | Nilotinib | 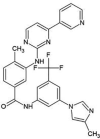 |
−9.4 | N4…..OD(Asn142) [3.03 Å] N1…..O(His164) [3.15 Å] |
Thr25, His41, Cys44, Met49, Phe140, Cys145, His163, Met165, Glu166 and Gln189 (N = 10) |
| ZINC000013831130 | Raltegravir |  |
−9.2 | O4…..O(His41) [2.95 Å] O2…..N(Cys145) [3.25 Å] N5…..OD1 (Asn142) [2.83 Å] |
Thr25, Thr26, Leu27, Cys44, Met49, Leu141, Gly143, Asp187, Arg188 and Gln189, (N = 10) |
| ZINC000003978005 | Dihydroergotamine | 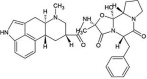 |
−9.2 | O4…..N(Ser144) [3.34 Å] O4…..N(Gly143) [2.67 Å] O1…..N(Gly143) [2.98 Å] |
Thr25, Leu27, His41, Cys44, Met49, Asn142, Cys145, His164, Met165, Glu166 and Gln189 (N = 11) |
| Control (G85) | SG85 |  |
−7.9 | O19…..N(Cys145) [3.20 Å] O19…..SG(Cys145) [3.28 Å] N49…..OE1(Gln189) [2.89 Å] O47…..N(Glu166) [3.02 Å] |
Thr25, Thr26, His41, Met49, Phe140, Leu141, Asn142, Gly143, Ser144, His163, His164, Met165, Asp187, Thr190, Ala191 and Gln192 (N = 16) |
