Fig 6. Mus81 and Rrm3 act in separate pathways to support replication.
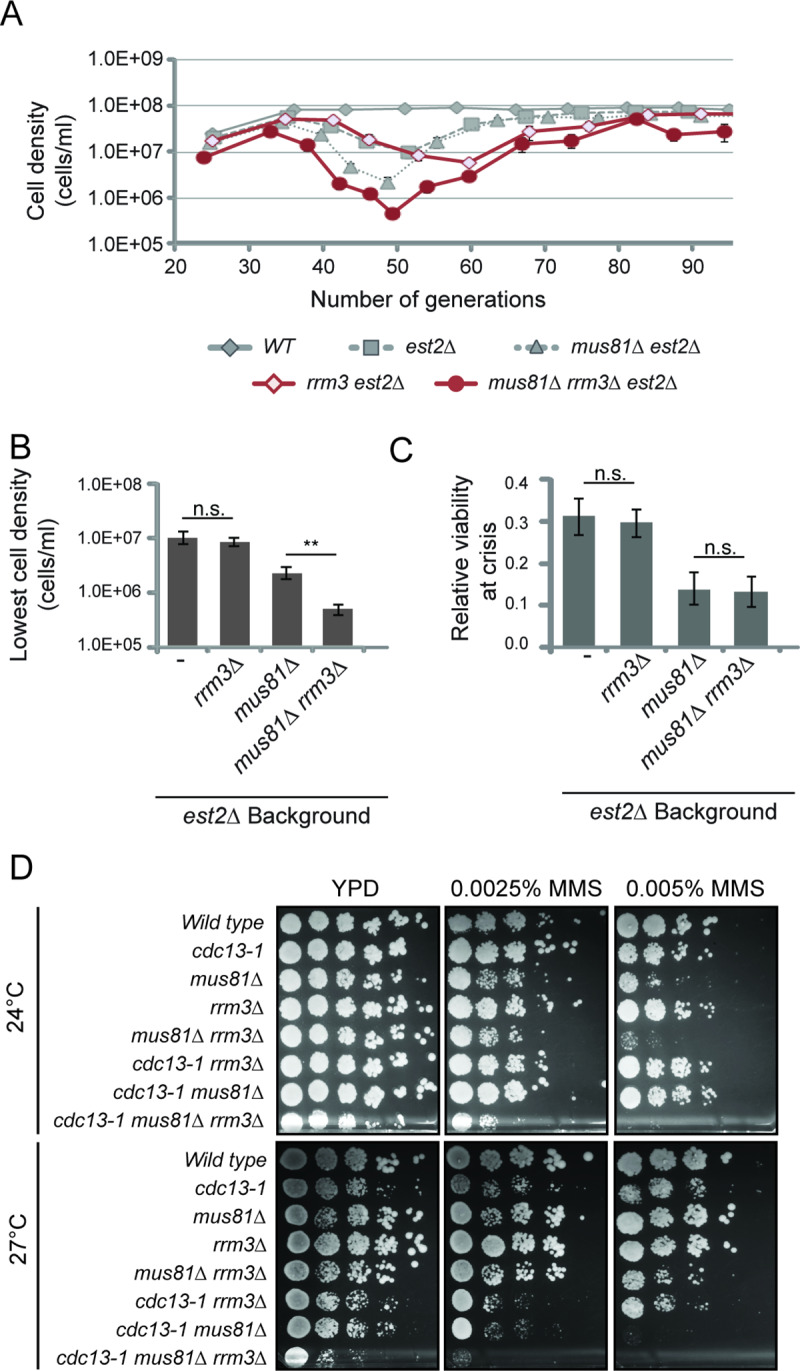
(A) Serial dilution assays monitoring cell density after 24 hours from an initial inoculate of 5x105 cells. Average cell density and one standard error is plotted for wild type (WT) (n = 20), est2Δ (n = 40), est2Δ mus81Δ (n = 27), est2Δ rrm3Δ (n = 9) and est2Δ mus81Δ rrm3Δ (n = 9) strains. Many error bars are smaller than the plotting symbol. (B) The lowest average cell density was plotted for all strains in A. (C) Relative cell viability was calculated from colony forming units at the generation with the lowest average cell density for the strains in (A). Statistical comparison of mean values to est2Δ single mutant. Student T-test is * P < 0.05, ** P < 0.01. Haploid strains est2Δ mus81Δ, est2Δ rrm3Δ were generated by sporulation of WDHY3653 as described in Materials and Methods. (D) Growth and viability was assessed in the presence and absence of the telomere capping protein Cdc13. Cells were chronically exposed to low doses of methyl methanesulfonate (MMS) or no additions as control. Cells were incubated at permissive (24°C) and semi-permissive (27°C) temperatures for two to four days depending on growth. Strains used for cdc13-1 genetics (C) are as follows: Wild type (W303-RAD5, MAT α), cdc13-1 (WDHY3086), mus81Δ (WDHY2601), rrm3Δ (WDHY3638), mus81Δ rrm3Δ (WDHY), and cdc13-1 mus81Δ (WDHY3058). Strains WDHY2601 and WDHY3661 were mated to create fresh haploids for cdc13-1 rrm3Δ and cdc13-1 mus81Δ rrm3Δ.
