Figure 1. F-actin nucleation determines dynamics and architecture of network assembly.
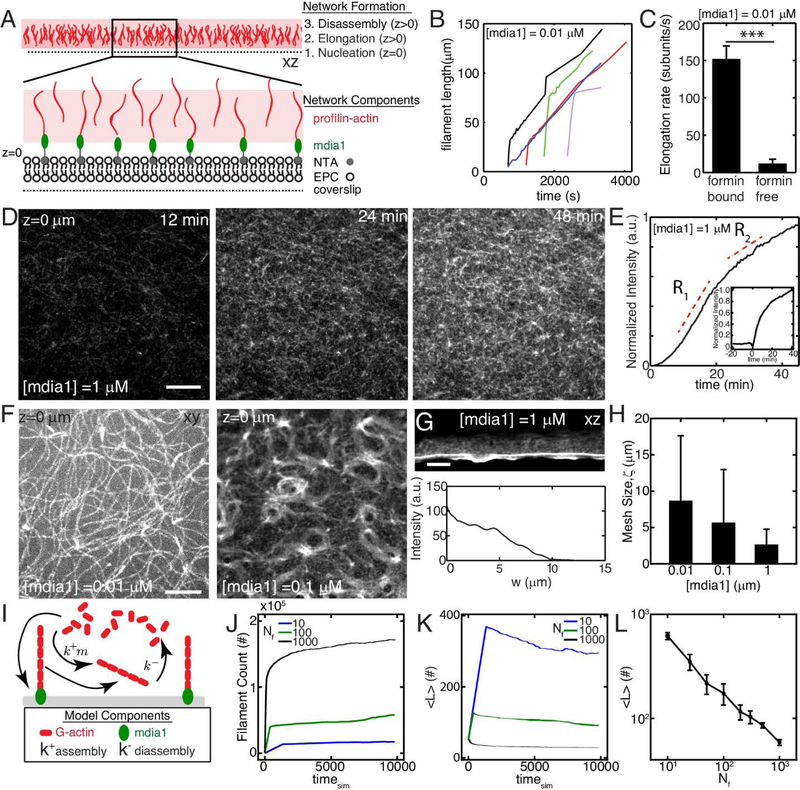
(A) Experimental setup where F-actin is nucleated from his-tagged mdia1 bound to NTA lipids within a model membrane. (B,C) Elongation rates of individual F-actin at 10 nM formin (N=4, 5, p<10−7). (D) F-actin fluorescence of network growth at 1 μM mdia1 concentration over time. (E) Normalized fluorescence intensity over time for the network in (D). (E,inset) Fluorescence intensity for a network where formin is added to proflin-actin at a later time (time=0 min). (F) Organization of F-actin for different concentrations of formin. (G) F-actin network nucleated from a microfluidic surface for a size view. (H) Mesh-size of the F-actin network as a function of formin concentration (N=3,4,3, p=ns) (I) Schematic of stochastic model for F-actin assembly from a limiting pool of G-actin. Model results for number of F-actin filaments (J), mean filament length (K), and equilibrium filament length for different number of nucleators (L). The pool size is 107 monomers for results in J-L. All scale bars are 10 μm. All error bars are mean ± 1 standard deviation.
