Fig. 1. Optimization of the detection of transcription.
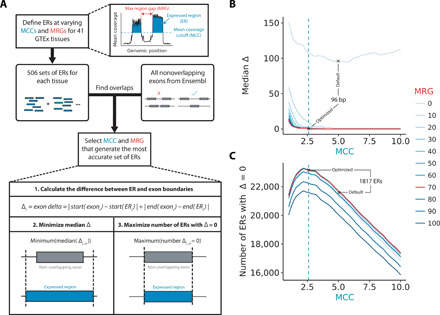
(A) Transcription in the form ERs was detected in an annotation-agnostic manner across 41 human tissues. The MCC is the number of reads supporting each base above which that base would be considered transcribed, and the MRG is the maximum number of bases between ERs below which adjacent ERs would be merged. MCC and MRG parameters were optimized for each tissue using the nonoverlapping exons from Ensembl v92 reference annotation. (B) Line plot illustrating the selection of the MCC and MRG that minimized the difference between ER and exon definitions (median exon delta). (C) Line plot illustrating the selection of the MCC and MRG that maximized the number of ERs that precisely matched exon definitions (exon delta = 0). The cerebellum tissue is plotted for (B) and (C), which is representative of the other GTEx tissues. Green and red lines indicate the optimal MCC (2.6) and MRG (70), respectively.
