Fig. 4. Unannotated ERs collectively serve an important function for humans, and a proportion can form potentially protein-coding transcripts.
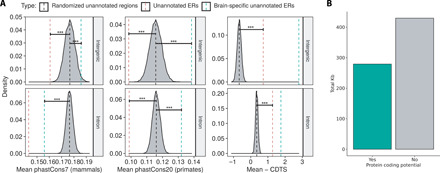
(A) Comparison of conservation (phastCons7/phastCons20) and constraint (CDTS) of intronic and intergenic ERs to 10,000 sets of random, length-matched intronic and intergenic regions. Unannotated ERs marked by the red dashed line are less conserved than expected by chance but are more constrained. Brain-specific ERs marked by the green dashed lines are among the most constrained. Data for the cerebellum shown and is representative of other GTEx tissues. ***P = <2 × 10–16. (B) The DNA sequence for ERs overlapping two junction reads was obtained and converted to amino acid sequence for all three possible frames. ERs (2168; 57%) lacked a stop codon in at least one frame and were considered potentially protein coding.
