Fig. 2. Clonal evolution in mCRC: From primary to metastasis and patient-derived xenograft.
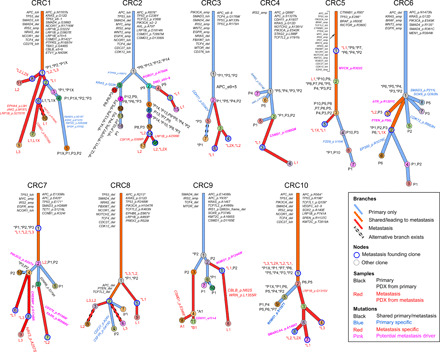
Each patient’s clonal history is presented by a tree whose nodes represent clones; branches represent evolution paths (length scaled by the square root of number of clonal marker mutations). Branches are labeled with potential driver mutations, and clone nodes are labeled with samples where the clones are found (with nonzero cellular fraction). A star (*) next to a sample indicates that the clone is the founding clone of the sample. Sample names are prefixed by a letter representing the site of tumor (P, primary; L, liver metastasis; A, abdominal wall metastasis; and B, brain metastasis). Suffix X indicates PDX. Clones marked as “alternative branch exists” were those predicted by ClonEvol to have an alternative position that does not change seeding model (see the Supplementary Figures of individual patients). Patient CRC11 was excluded from clonal evolution analysis because of low-quality primary sample.
