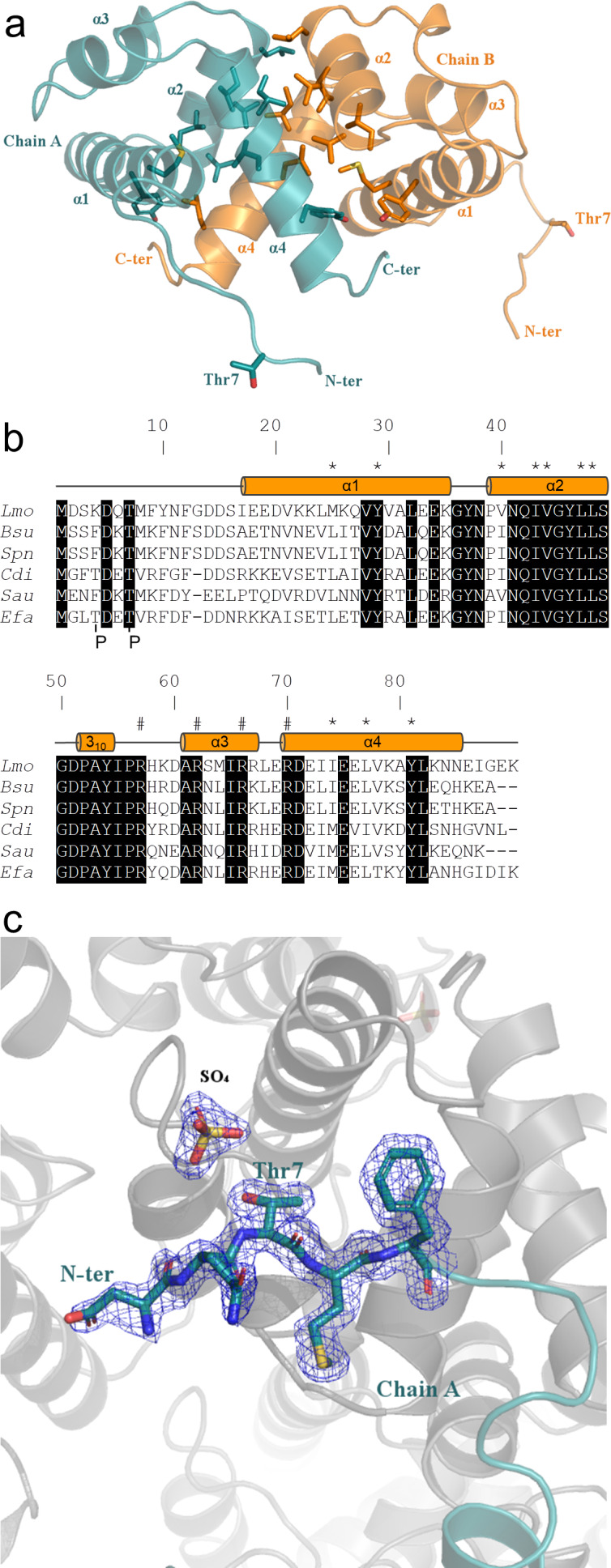
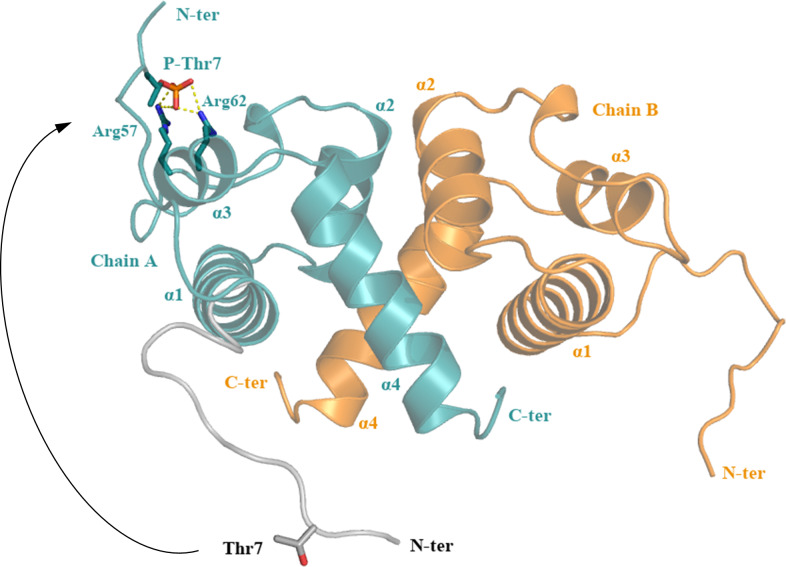
Figure 6. Crystal structure of ReoM.
(A) The structure of ReoM depicted as a cartoon with each protomer in the dimer coloured separately (cyan and orange). The secondary structure elements are numbered according to their position in the amino acid sequence. Thr7 and some of the key amino acids in the dimer interface and the hydrophobic core are drawn as stick figures. (B) Sequence alignment of ReoM (Lmo) and its homologues from Bacillus subtilis (Bsu), Streptococcus pneumoniae (Spn), Clostridium difficile (Cdi) and Staphylococcus aureus (Sau) with the sequence of IreB from Enterococcus faecalis (Efa) underneath. Amino acid sequence numbers pertain to ReoM and the site of phosphorylation in ReoM (Thr7) and the twin phosphorylations in IreB (minor site: Thr4; major site: Thr7) are highlighted. Invariant amino acids are shaded black, residues in the ReoM dimer interface have an asterisk above, and the secondary structure elements are defined by cylinders above the alignment. Arginine residues mutated in this study are indicated by a hashtag above the alignment. (C) The final 2Fobs-Fcalc electron density map, contoured at a level of 0.42 e-/Å3, of the N-terminal region in the immediate vicinity of Thr7 in chain A of the ReoM dimer indicates that the protein model could be built with confidence even though this region contains no secondary structure elements.