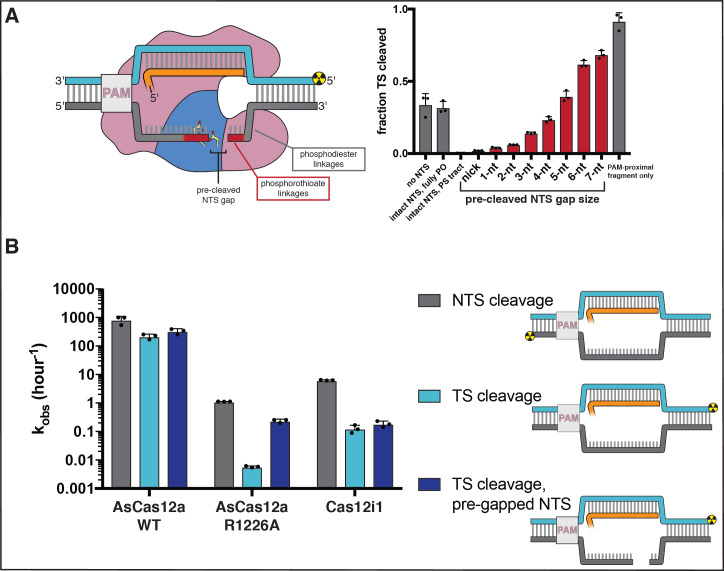
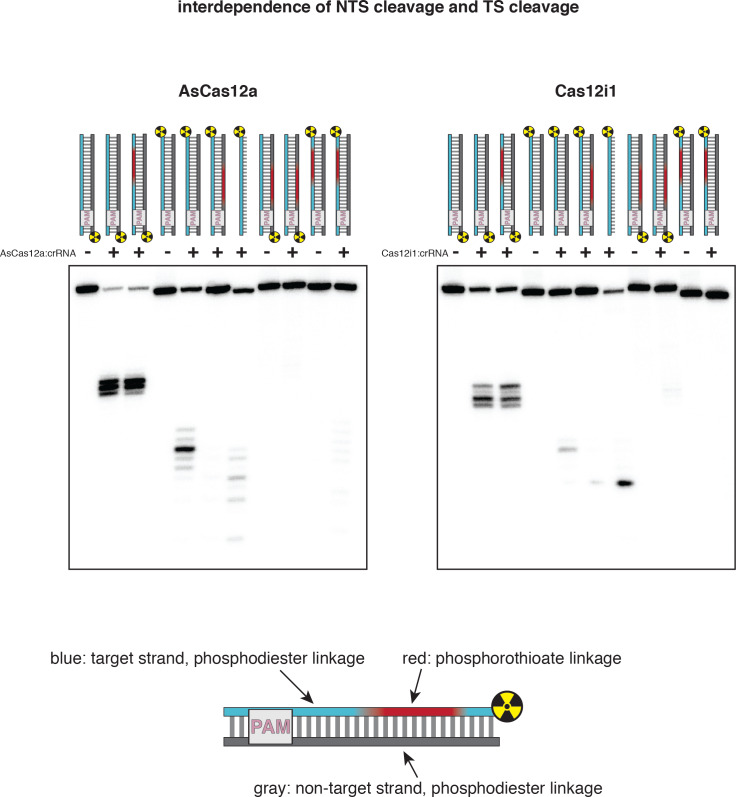
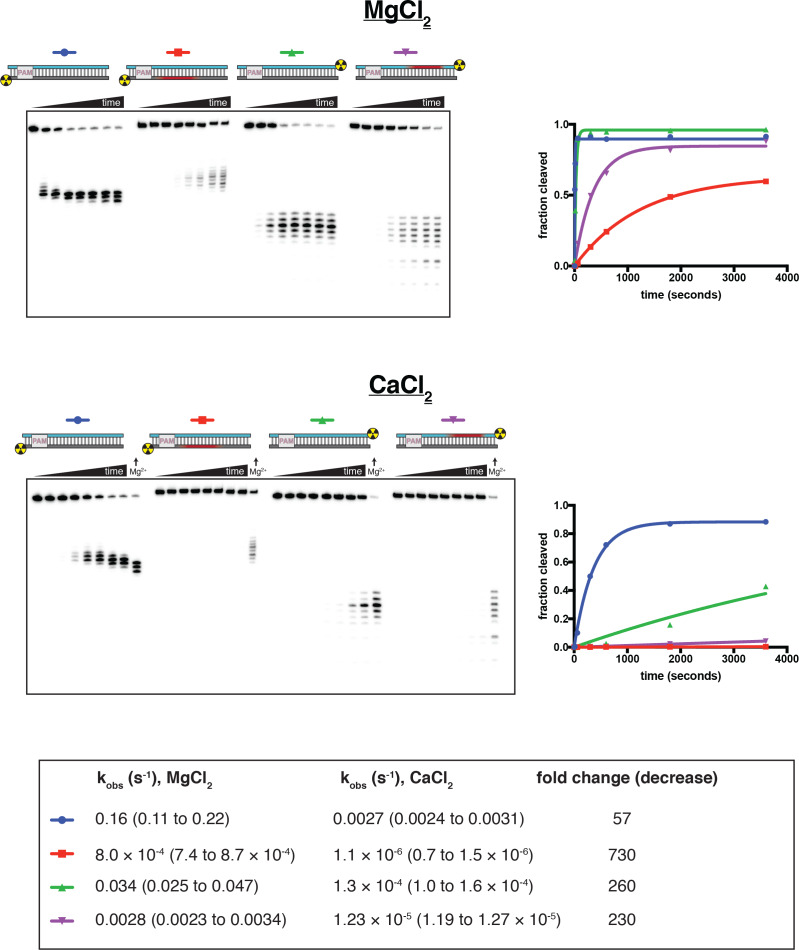
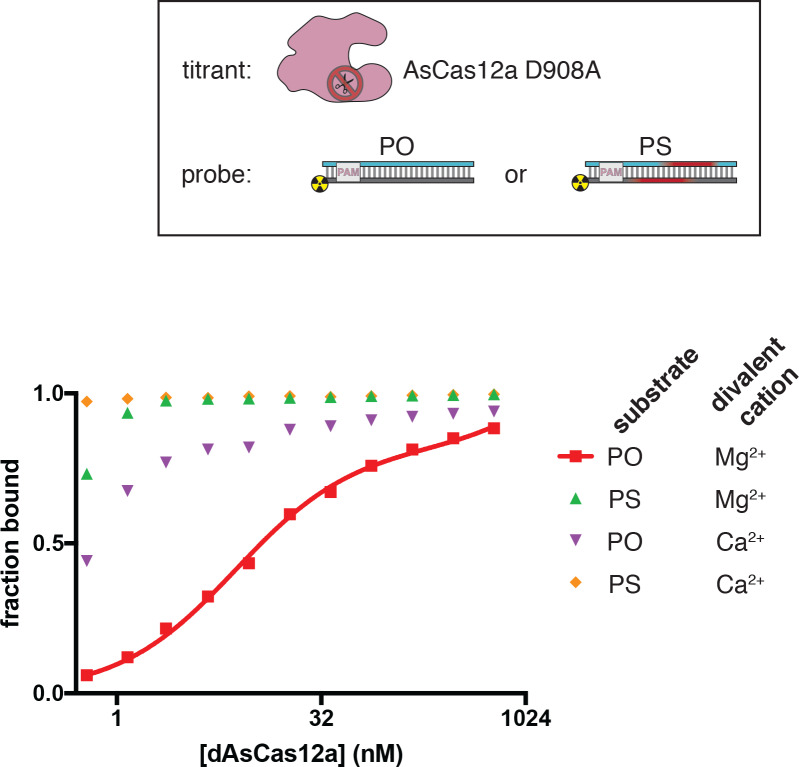
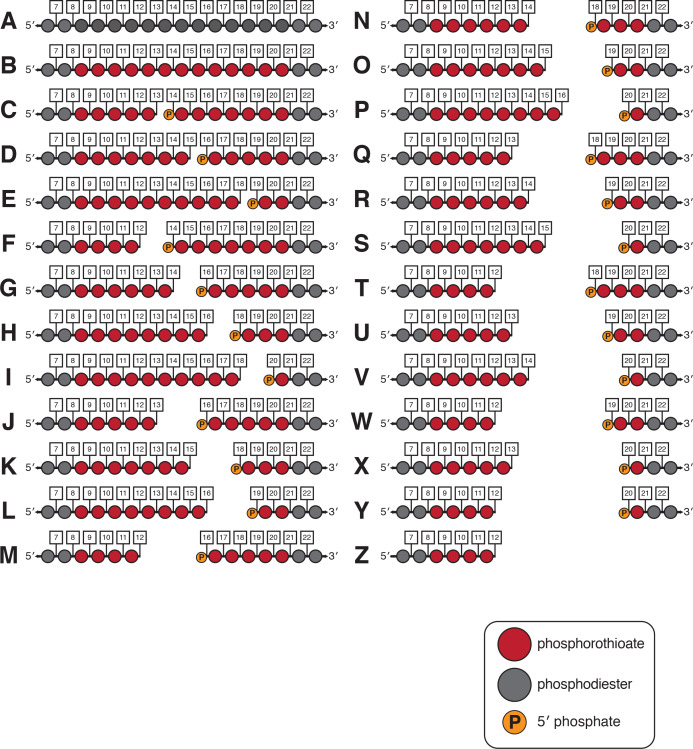
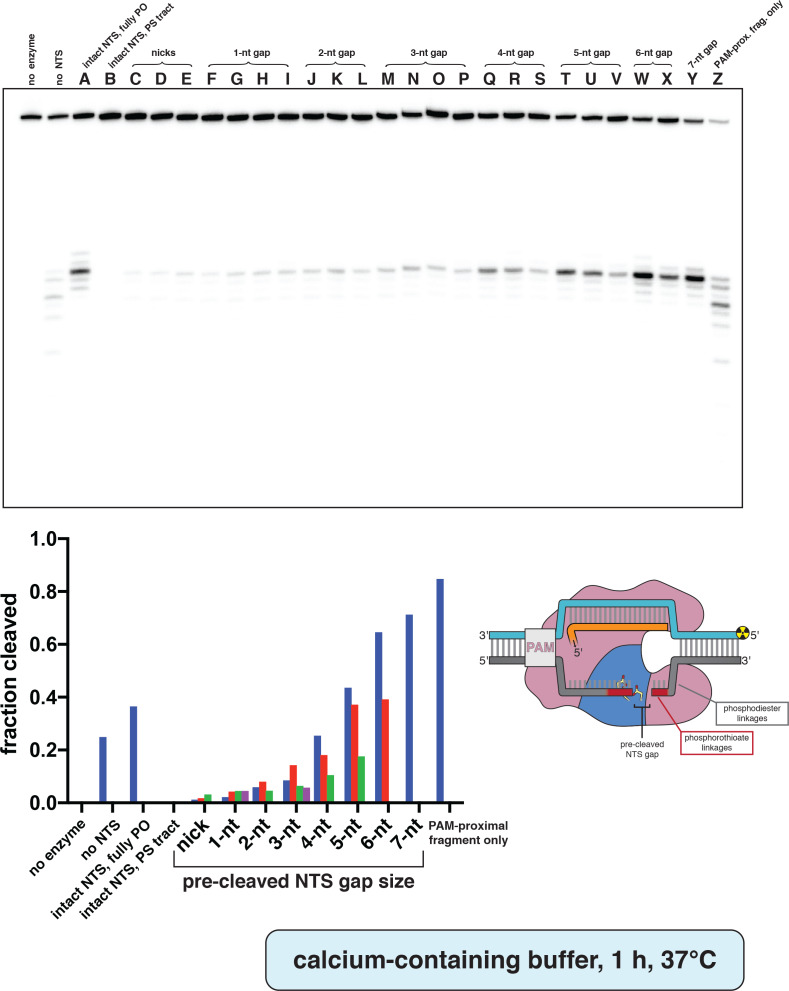
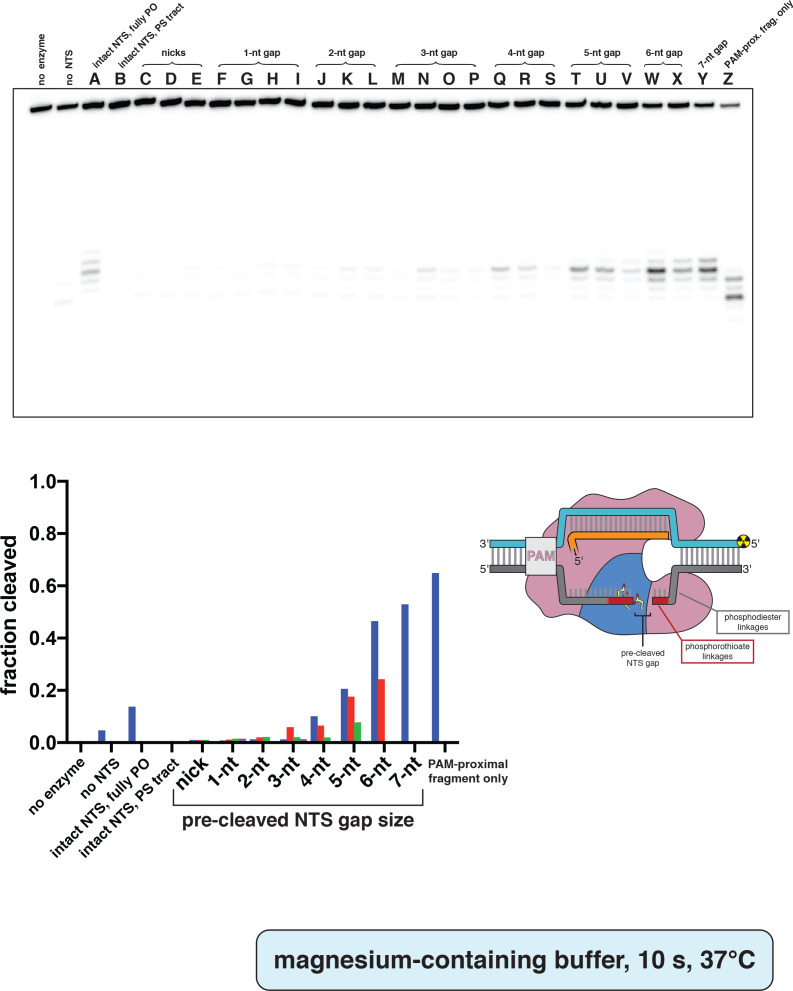
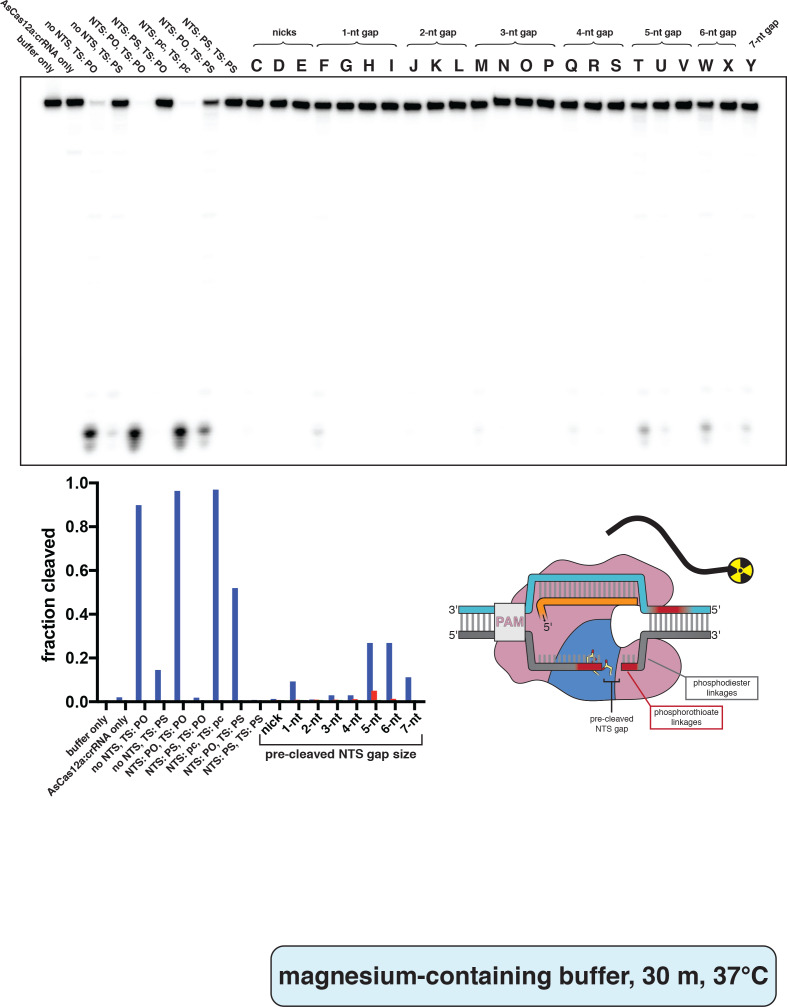
Appendix 2—figure 2. Non-target-strand gap formation poses a kinetic barrier to target-strand cleavage for AsCas12a.
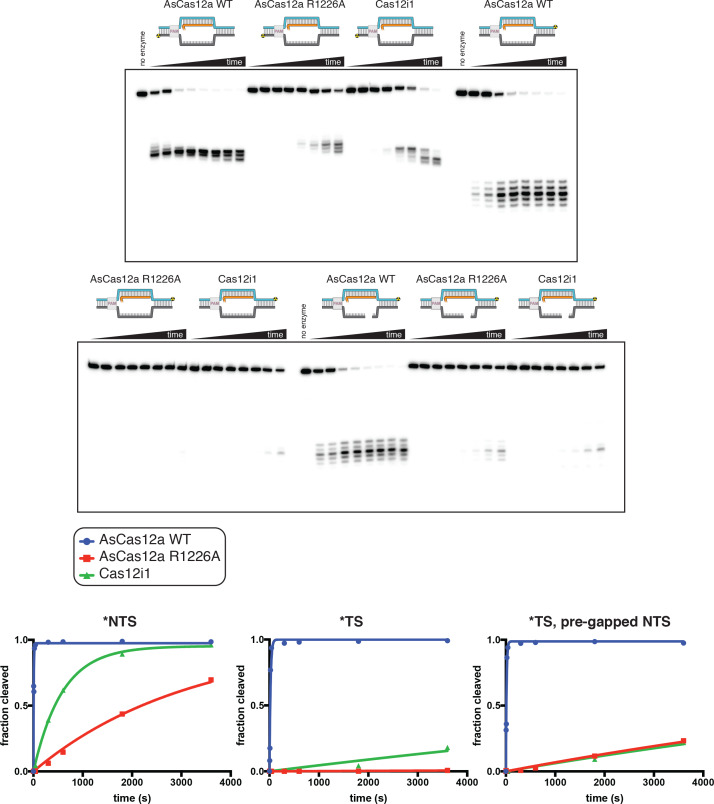
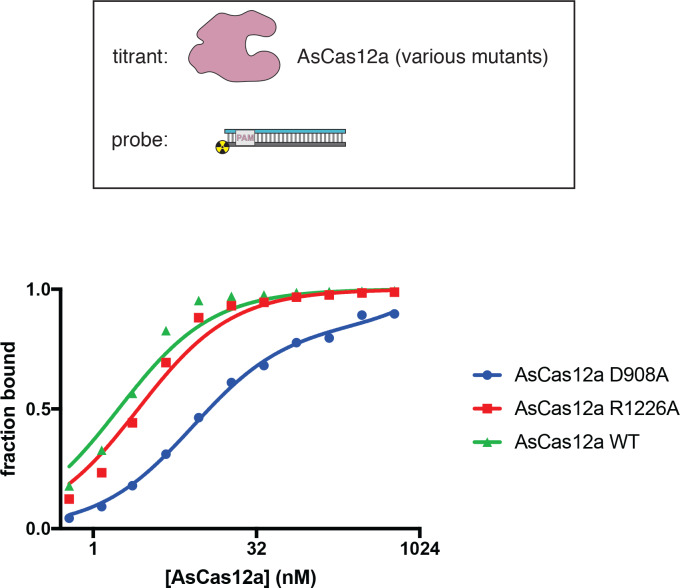
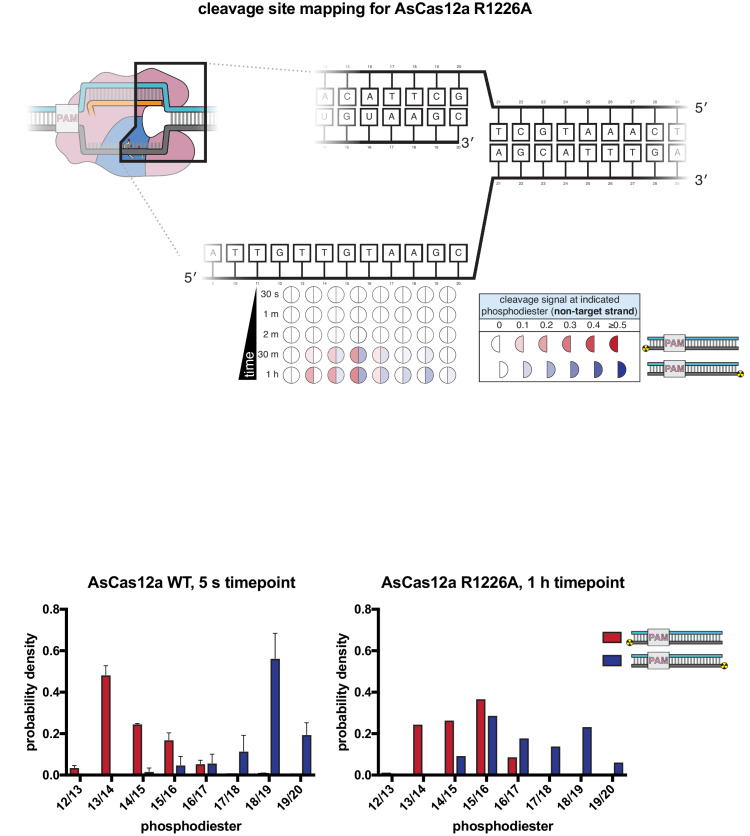
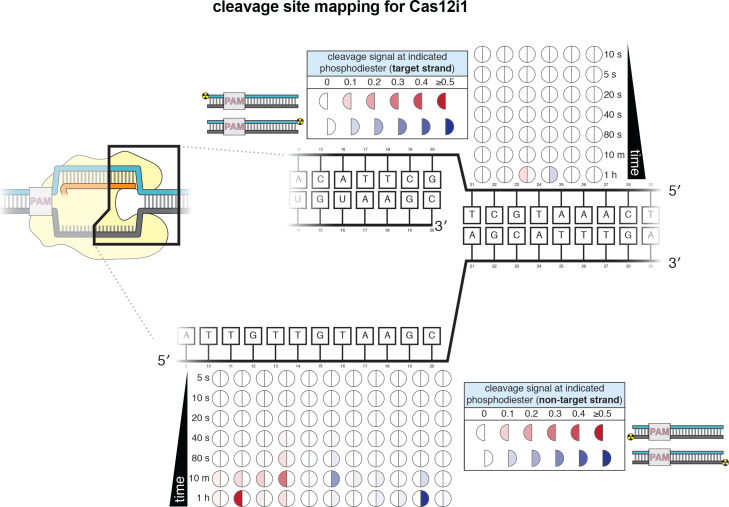
(A) Extent of target-strand cleavage by wild type AsCas12a in the presence of various non-target-strand variants, as resolved by denaturing PAGE (phosphorimage in Appendix 2—figure 2—figure supplement 5). Cas12a surveillance complex (100 nM AsCas12a, 120 nM crRNA) was added to 1 nM pre-hybridized target DNA radiolabeled on the 5' end of the TS and allowed to incubate in cleavage buffer with 5 mM CaCl2 for 1 hr at 37°C prior to quenching. In the schematic, the red portion of the NTS denotes phosphorothioate (PS) linkages; the gray portion denotes phosphodiester (PO) linkages. In the graph, red bars denote reactions with a PS-containing NTS variant; gray bars denote reactions either with no NTS or with an NTS variant containing only PO-linkages. From left to right (omitting the no-NTS control), the NTS variants used were A, B, D, G, J, N, Q, T, W, Y, Z, as schematized in Appendix 2—figure 2—figure supplement 4. Columns and associated error bars indicate the mean and standard deviation of three replicates. (B) Cleavage kinetics of NTS, TS, and TS complexed with a pre-gapped NTS (NTS contains a 5-nt gap). 100 nM protein and 120 nM cognate crRNA were incubated with 2 nM DNA target with a 5' radiolabel on the indicated strand at 37°C for various timepoints, followed by quenching and resolution by denaturing PAGE. Representative phosphorimages and quantifications are shown in Appendix 2—figure 2—figure supplement 8. Columns and associated error bars indicate the mean and standard deviation of three replicates.