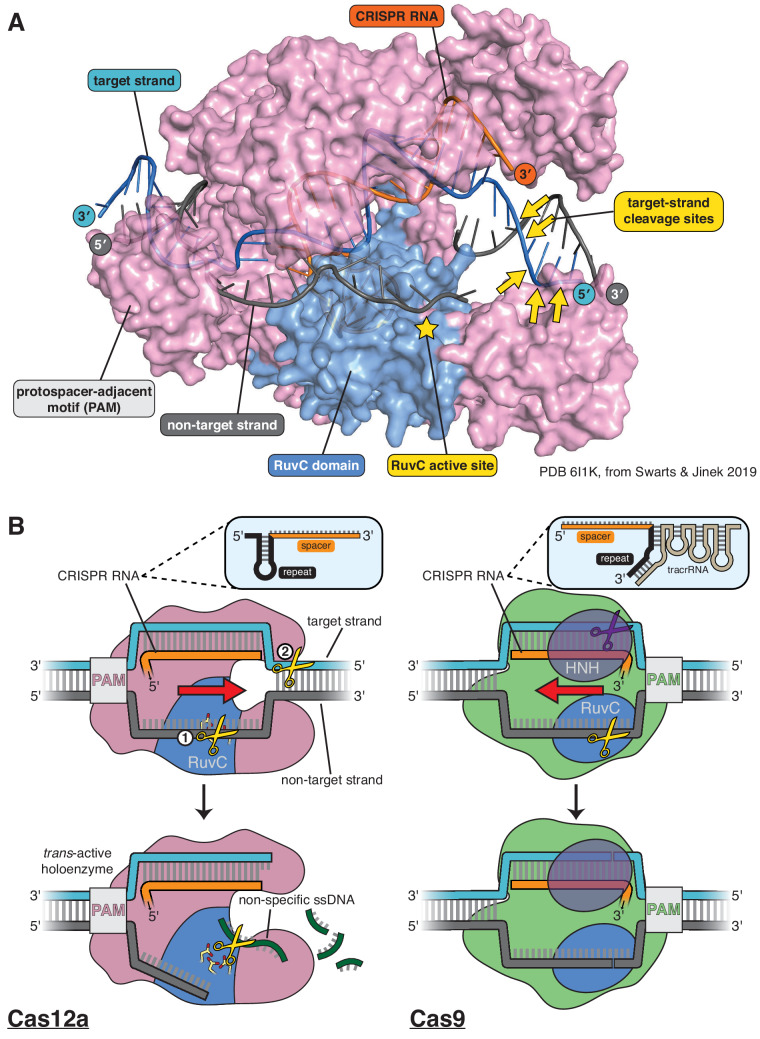
Figure 1. Structure of Cas12a and comparison of its DNA cleavage pathway to that of Cas9.
(A) Crystal structure of the DNA-bound Cas12a interference complex from Francisella novicida (FnCas12a, PDB 61IK) (Swarts and Jinek, 2019). While the protein ortholog used for most experiments in this manuscript is from Acidaminoccus species (AsCas12a,~40% identity to FnCas12a), the FnCas12a crystal structure shown here represents the most complete structure of such a complex to date, most notably with respect to the DNA at the target-strand cleavage sites. We did not perform any experiments with the particular DNA sequence used by Swarts and Jinek in crystallization, so the scissile phosphodiesters indicated were determined for a different sequence (see Appendix 2—figure 1, Appendix 2—figure 1—figure supplement 6) and superimposed onto the structural model according to their distance from the PAM (in terms of number of nucleotides). The discontinuity modeled into the non-target strand corresponds to positions of weak electron density in the crystal structure, which could have been due to some combination of disorder of the (intact) intervening tract and/or in crystallo hydrolysis and dissociation of the intervening tract. (B) For Cas12a, successful R-loop formation results in activation of the RuvC DNase active site to cleave three classes of DNA substrates (yellow scissors): the non-target strand (in cis), the target strand (in cis), and non-specific ssDNA (in trans). Circled numbers indicate the required order of cis strand cleavage; three conserved active site carboxylates of the RuvC DNase are shown in yellow and red; ‘PAM’ indicates the protospacer-adjacent motif; red arrow indicates the direction in which the R-loop is opened. Cas9 contains two DNase domains: the RuvC domain cleaves the non-target strand, and the HNH domain cleaves the target strand.