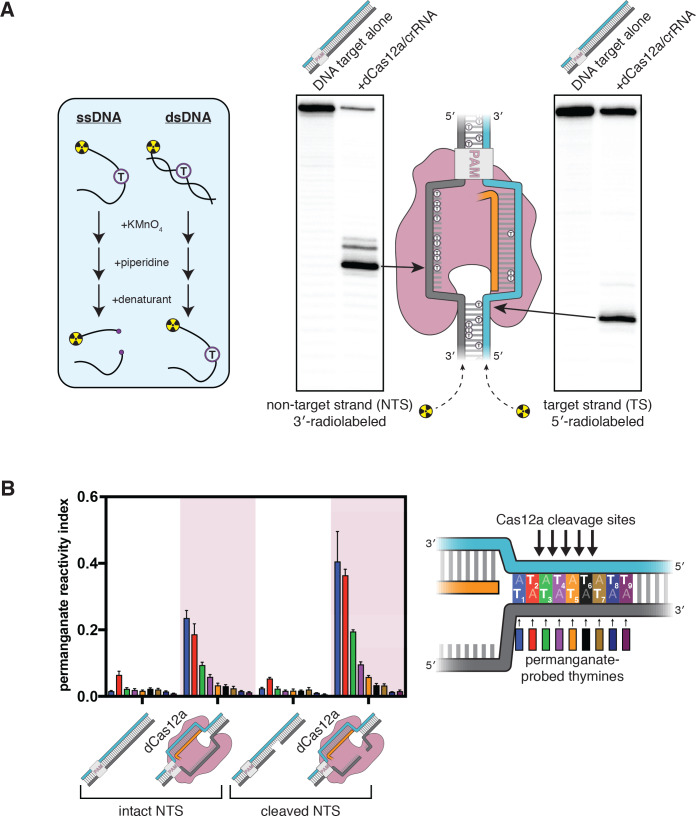
Figure 2. The target-strand cleavage site becomes distorted upon R-loop formation.
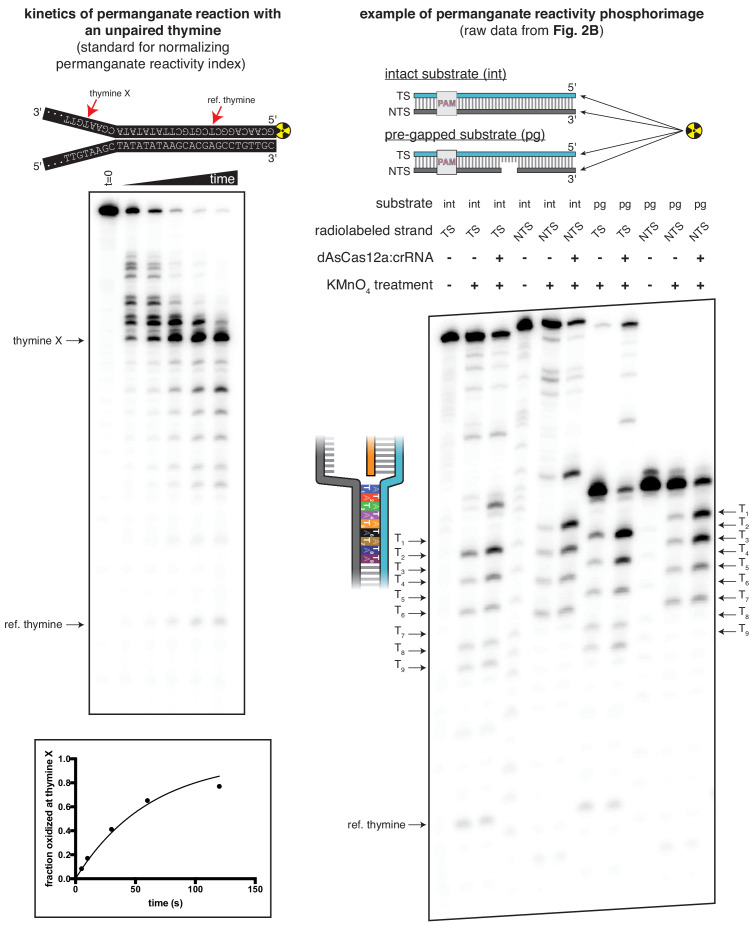
(A) Denaturing PAGE phosphorimages of piperidine-treated permanganate oxidation products, demonstrating the assay’s ability to detect non-B-form DNA conformations within and adjacent to a dCas12a-generated R-loop. Permanganate reactions were quenched after 10 s at 30°C. Each thymine in the DNA substrate is shown as a circled T. (B) Permanganate reactivity of a PAM-distal R-loop flank whose sequence was changed (as compared to the native protospacer sequence that was probed in A) to contain more thymines, with an intact or cleaved non-target strand (‘cleaved NTS’ indicates that there is a 5-nt gap in the NTS—see Appendix 2). Permanganate reactions were quenched after 2 min at 30°C. A raw phosphorimage is shown in Figure 2—figure supplement 3. The permanganate reactivity index (PRI) is an estimate of the rate of oxidation at each thymine, normalized such that PRI = 1 for a fully single-stranded thymine (see Materials and methods). Columns and associated error bars indicate the mean and standard deviation of three replicates. The phosphodiester bonds normally cleaved by WT Cas12a are indicated with arrows on the substrate schematic for reference, but note that the complexes being probed with permanganate were formed with dCas12a.