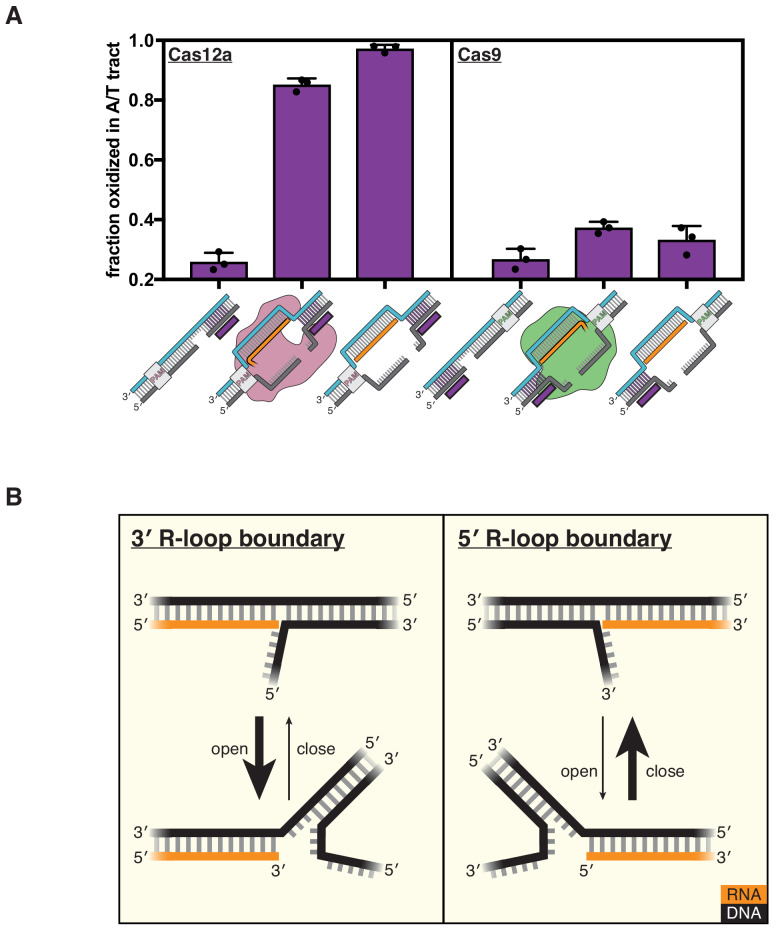
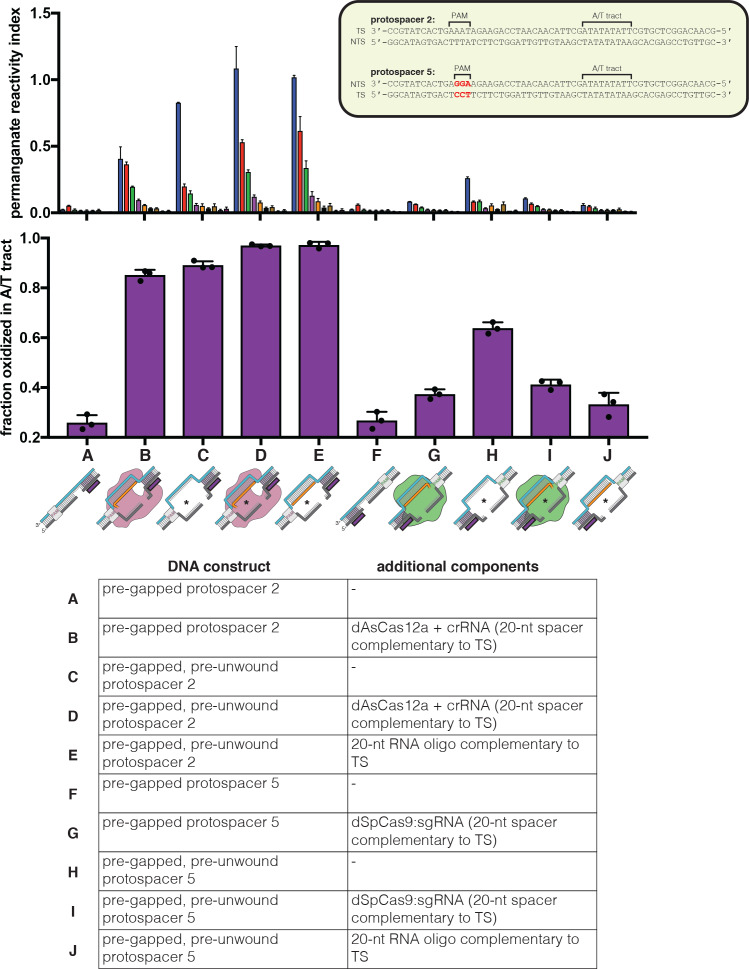
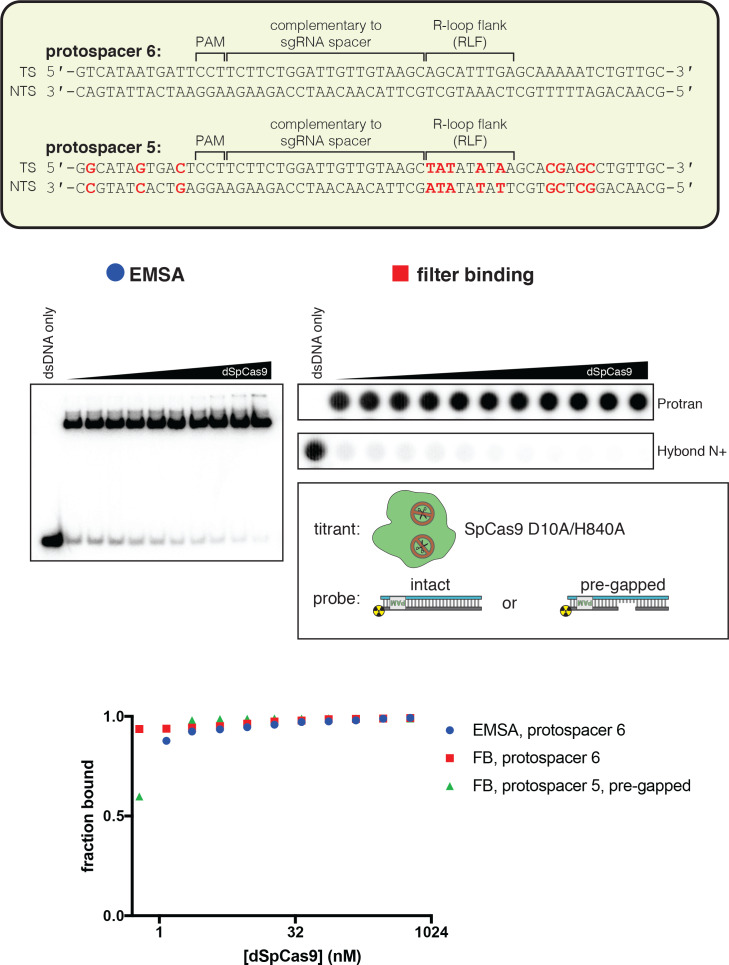
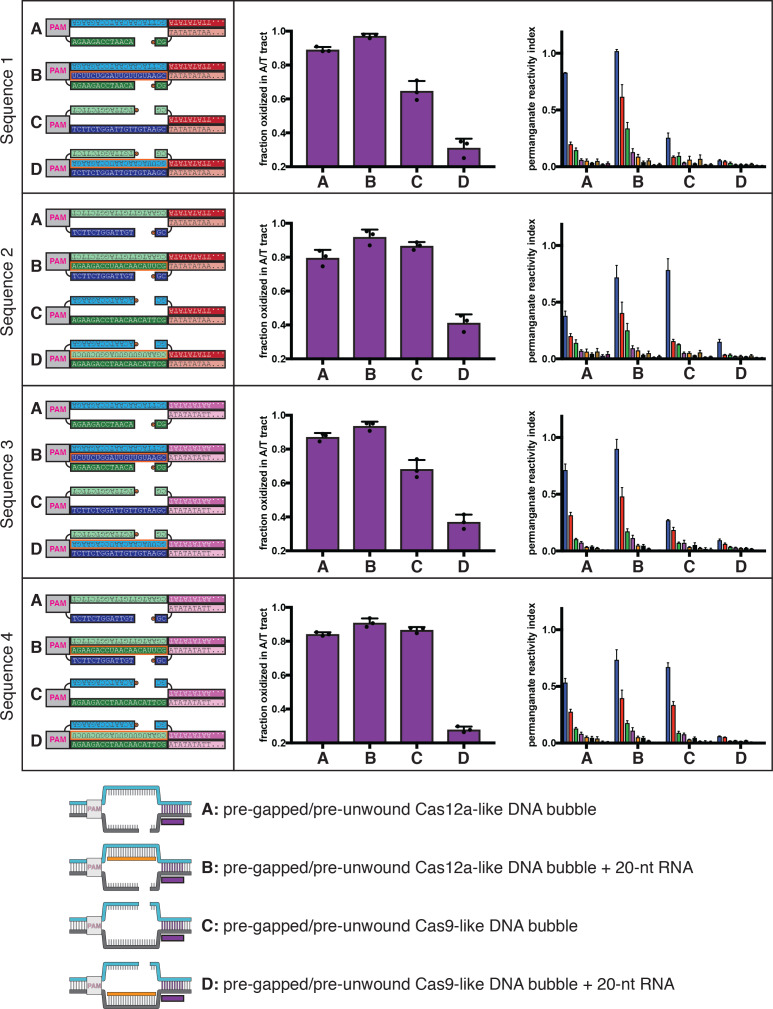
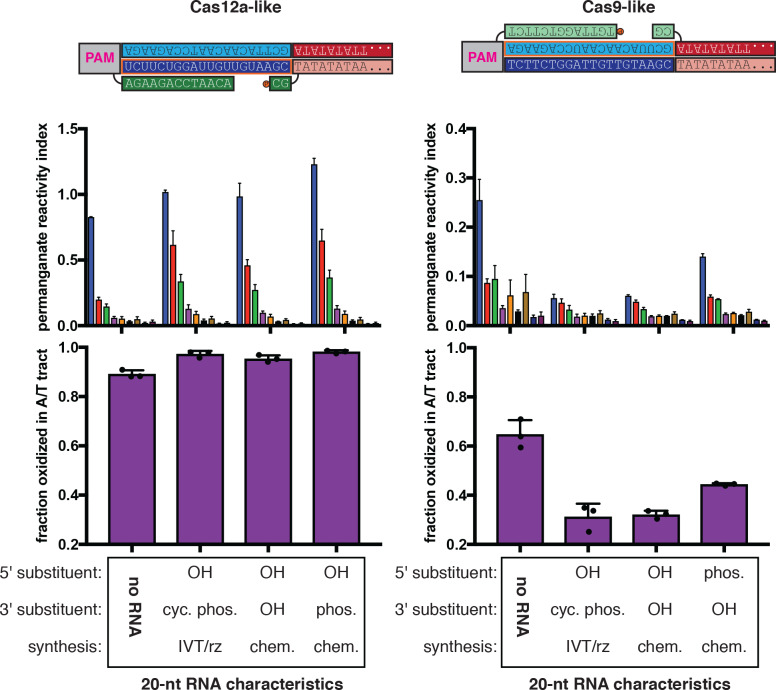
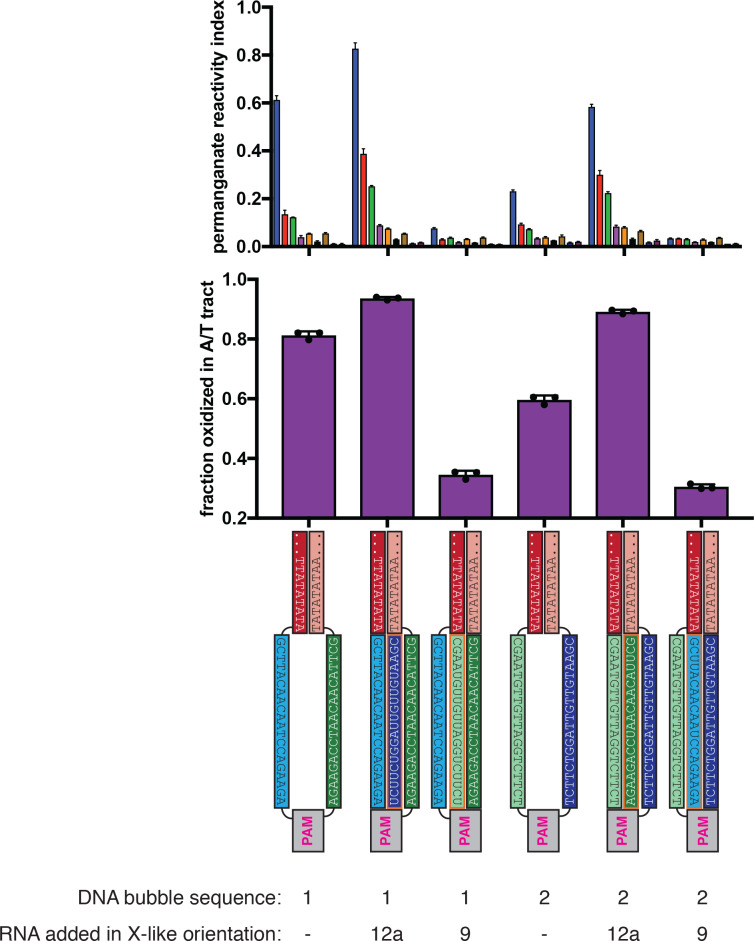
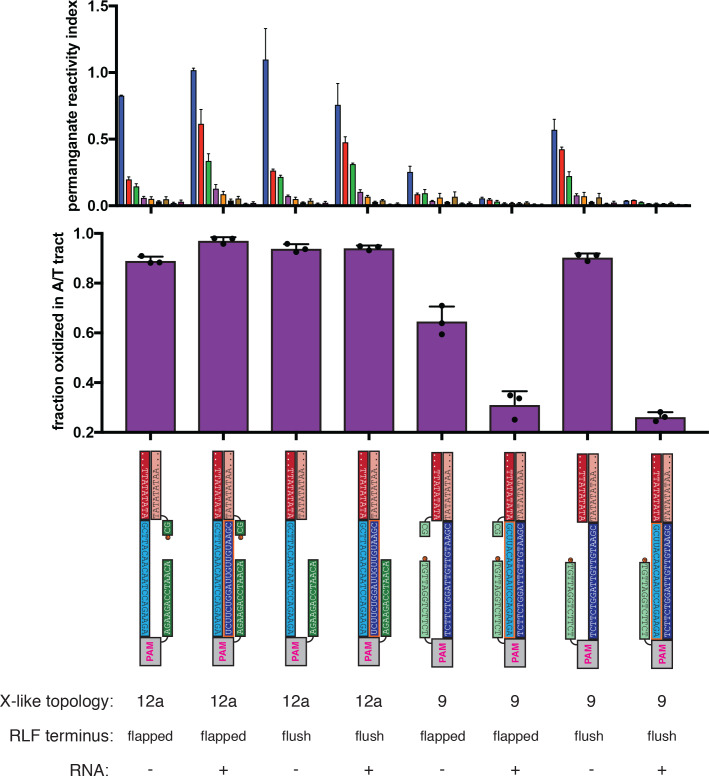
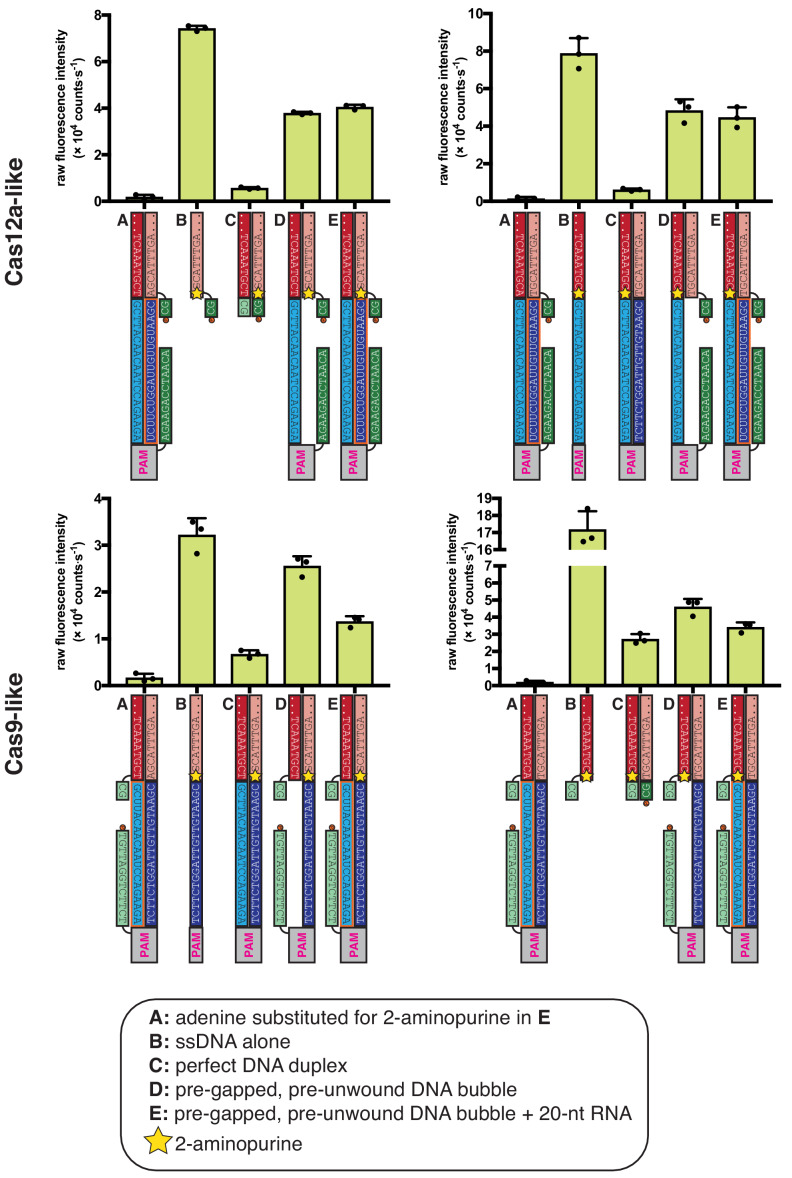
Figure 4. DNA distortion is protein-independent and unique to 3' R-loop flanks.
(A) Permanganate reactivity of the A/T tract in a dCas12a R-loop, a dCas9 R-loop, and their protein-free mimics. The y-axis denotes the fraction of DNA molecules estimated to have been oxidized on at least one thymine within the A/T tract (see Materials and methods). Purple rectangles alongside DNA schematics indicate the location of the tract of DNA whose permanganate reactivity is being quantified. Columns and associated error bars indicate the mean and standard deviation of three replicates. (B) Model for the relative conformational dynamics of 3' and 5' R-loop boundaries, as suggested by permanganate reactivity experiments. The depth of fraying shown (three base pairs) was chosen arbitrarily for the schematic and should not be interpreted as a uniquely stable ‘open’ structure (see Materials and methods).