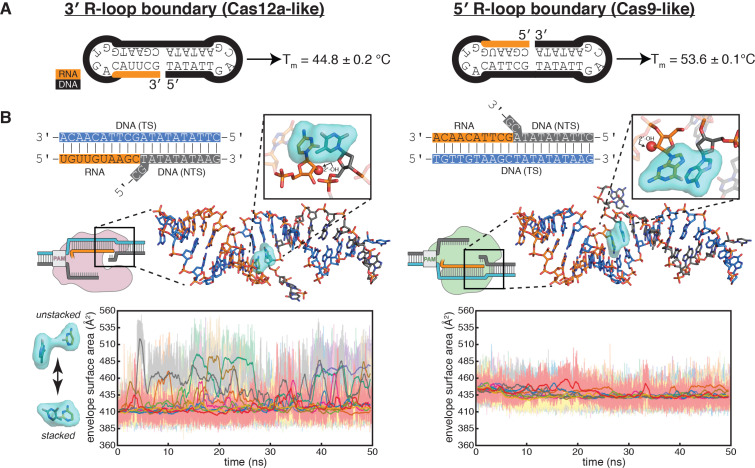
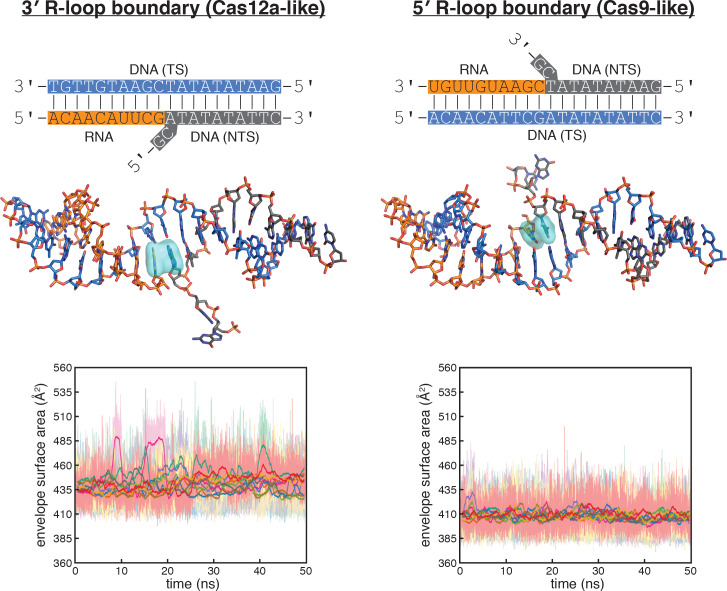
Figure 5. Energetics of base stacking at the R-loop boundary probed by optical measurements and molecular dynamics simulations.
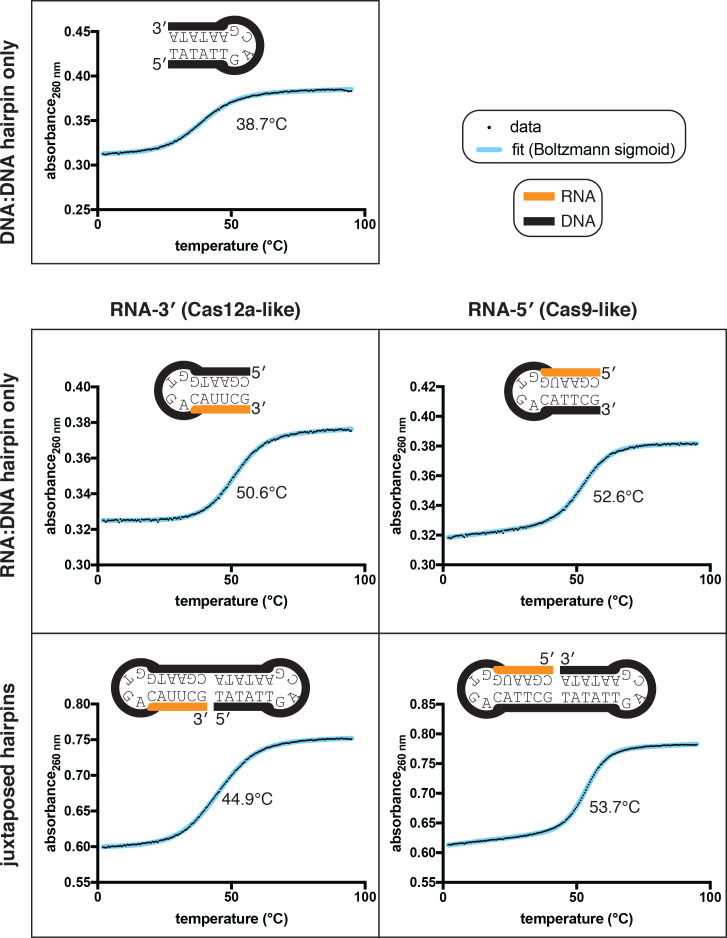
(A) Melting temperatures of nicked-dumbbell constructs that recapitulate each type of R-loop boundary, determined by monitoring absorbance of ultraviolet light while slowly cooling samples from 95°C to 2°C. Reported values show mean and standard deviation of three replicates. See Figure 5—figure supplement 1 for refolding curves and control constructs. (B) Molecular dynamics simulations reveal nucleobase unstacking in 3' R-loop boundaries but not in 5' R-loop boundaries. At the top left is a schematized version of the true structural model shown immediately below (this coaxially stacked conformation is the starting structure that was used for simulation); hydrogens were present in the simulated model and analyses but are omitted from representations here for clarity. The simulated model contained only the nucleic acid molecules shown in stick representation; the protein and remainder of the R-loop are drawn in a schematic only to orient the reader as to where the simulated structure would fit into a full DNA-bound CRISPR interference complex; the Cas9-orientation R-loop is drawn with a Cas12a-like NTS gap to reflect the simulated model. The inset is a closeup of the two nucleotides on the ‘flapped’ side of the junction in the structural model; the 2'-OH is shown as a red sphere. Envelope surface area (ESA) was determined by isolating two nucleobases of the interhelical stack—that on the RNA terminus and that stacked upon it from the NTS—and calculating the surface area of the volume they jointly occupy over the course of each trajectory (envelope shown in cyan). High ESA values reflect unstacking of nucleobases, whereas low ESA values reflect a stacked architecture similar to that of the starting conformation. Pale lines are absolute ESA values, and bold lines are moving averages (1-ns sliding window). Data from ten independent 50-ns trajectories are shown in different colors. Simulations of a second set of sequences are described in Figure 5—figure supplement 2.