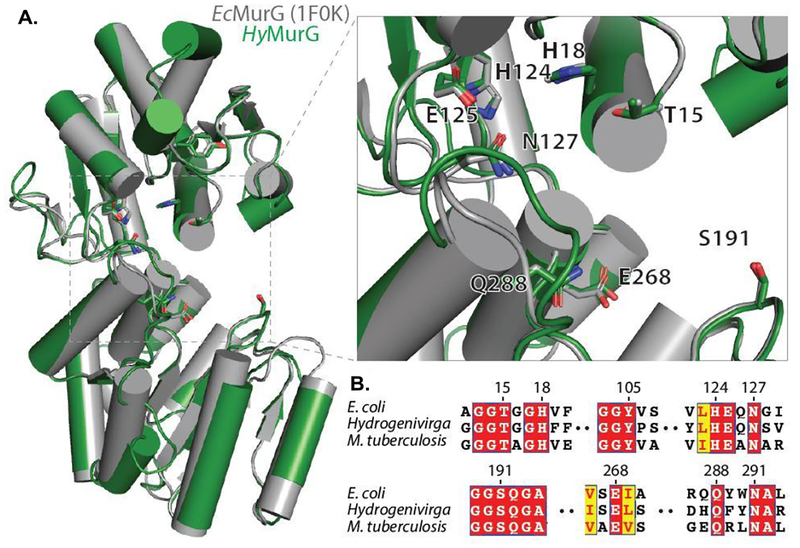
Figure 10.
Conservation of MurG homologs. A: Structural models of MurG with E. coli (PDB ID: 1F0K) aligned to a homology model of MurG from Hydrogenivirga sp. HyMurG colored in green and EcMurG in gray. Putative active site is magnified in the inset with important catalytic residues as sticks, numbering based on EcMurG. Homology model was created in SWISS-MODEL [Waterhouse et al. 201858 using EcMurG (PDB ID: 1F0K) chain B as a template. Figure was generated using PyMOL (Version 2.2.3, Schrödinger, LLC, Portland, OR, USA)59. B: Alignment of MurG sequences from EcMurG, HyMurG, and MtbMurG highlighting residues from A. Alignment made using T-Coffee and ESPript [Tommaso et al. 2011; Robert et al. 2014]60,61.